High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates
- PMID: 12147485
- PMCID: PMC124033
- DOI: 10.1128/AEM.68.8.3878-3885.2002
High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates
Abstract
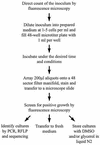
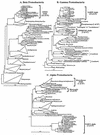
Microbial diversity studies based on the cloning and sequencing of DNA from nature support the conclusion that only a fraction of the microbial diversity is currently represented in culture collections. Out of over 40 known prokaryotic phyla, only half have cultured representatives. In an effort to culture the uncultured phylotypes from oligotrophic marine ecosystems, we developed high-throughput culturing procedures that utilize the concept of extinction culturing to isolate cultures in small volumes of low-nutrient media. In these experiments, marine bacteria were isolated and cultivated at in situ substrate concentrations-typically 3 orders of magnitude less than common laboratory media. Microtiter plates and a newly developed procedure for making cell arrays were employed to raise the throughput rate and lower detection sensitivity, permitting cell enumeration from 200-microl aliquots of cultures with densities as low as 10(3) cells/ml. Approximately 2,500 extinction cultures from 11 separate samplings of marine bacterioplankton were screened over the course of 3 years. Up to 14% of the cells collected from coastal seawater were cultured by this method, which was 14- to 1,400-fold higher than the numbers obtained by traditional microbiological culturing techniques. Among the microorganisms cultured were four unique cell lineages that belong to previously uncultured or undescribed marine Proteobacteria clades known from environmental gene cloning studies. These cultures are related to the clades SAR11 (alpha subclass), OM43 (beta subclass), SAR92 (gamma subclass), and OM60/OM241 (gamma subclass). This method proved successful for the cultivation of previously uncultured marine bacterioplankton that have consistently been found in marine clone libraries.
Figures

References
-
- Ammerman, J. W., J. A. Fuhrman, Å. Hagström, and F. Azam. 1984. Bacterioplankton growth in seawater. I. Growth kinetics and cellular characteristics in seawater cultures. Mar. Ecol. Prog. Ser. 18:31-39.
-
- Béjà, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. B. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902-1906. - PubMed
-
- Béjà, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. DeLong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2:516-529. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

