Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea
- PMID: 12147499
- PMCID: PMC124024
- DOI: 10.1128/AEM.68.8.3978-3987.2002
Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea
Abstract
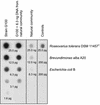
The effect of signal molecules on the cultivation efficiency of bacteria from the Gotland Deep in the central Baltic Sea was investigated. Numbers of cultivated cells were determined by the most-probable-number (MPN) technique. Artificial brackish water supplemented with different carbon substrates at low concentrations (200 microM each) was employed as the growth medium. Compared to the results of previous studies, this approach yielded significantly higher cultivation efficiencies (up to 11% in fluid media). A further and pronounced increase in cultivation success was accomplished by the addition of cyclic AMP (cAMP), N-butyryl homoserine lactone, or N-oxohexanoyl-DL-homoserine lactone at a low concentration of 10 microM. The most effective inducer was cAMP, which led to cultivation efficiencies of up to 100% of total bacterial counts. From the highest positive dilutions of these latter MPN series, several strains were isolated in pure culture and one strain (G100) was used to study the physiological effect of cAMP. Dot blot hybridization revealed, however, that strain G100 represented only a small fraction of the total bacterial community. This points towards an inherent limitation of the MPN approach, which does not necessarily recover abundant species from highly diverse communities. Bacterial cells of strain G100 that were starved for 6 weeks attained a higher growth rate and a higher biomass yield when resuscitated in the presence of cAMP instead of AMP.
Figures




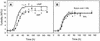
References
-
- Ammerman, J. W., and F. Azam. 1981. Dissolved cyclic adenosine monophosphate (cAMP) in the sea and uptake of cAMP by marine bacteria. Mar. Ecol. Prog. Ser. 5:85-89.
-
- Babenzien, H. D., and H. Sass. 1999. Desulfurikation, p. 435-444. In W. von Tümpling and G. Friedrich (ed.), Biologische Gewässeruntersuchung, vol. 2. Gustav Fischer Verlag, Jena, Germany.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

