Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR
- PMID: 12149343
- PMCID: PMC120665
- DOI: 10.1128/JCM.40.8.2860-2865.2002
Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR
Abstract
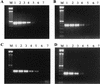
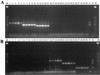
A multiplex PCR method was developed to identify simultaneously multiple fungal pathogens in a single reaction. Five sets of species-specific primers were designed from the internal transcribed spacer (ITS) regions, ITS1 and ITS2, of the rRNA gene to identify Candida albicans, Candida glabrata, Candida parapsilosis, Candida tropicalis, and Aspergillus fumigatus. Another set of previously published ITS primers, CN4 and CN5, were used to identify Cryptococcus neoformans. Three sets of primers were used in one multiplex PCR to identify three different species. Six different species of pathogenic fungi can be identified with two multiplex PCRs. Furthermore, instead of using templates of purified genomic DNA, we performed the PCR directly from yeast colonies or cultures, which simplified the procedure and precluded contamination during the extraction of DNA. A total of 242 fungal isolates were tested, representing 13 species of yeasts, four species of Aspergillus, and three zygomycetes. The multiplex PCR was tested on isolated DNA or fungal colonies, and both provided 100% sensitivity and specificity. However, DNA from only about half the molds could be amplified directly from mycelial fragments, while DNA from every yeast colony was amplified. This multiplex PCR method provides a rapid, simple, and reliable alternative to conventional methods to identify common clinical fungal isolates.
Figures

Similar articles
-
Rapid identification of fungal pathogens in BacT/ALERT, BACTEC, and BBL MGIT media using polymerase chain reaction and DNA sequencing of the internal transcribed spacer regions.Diagn Microbiol Infect Dis. 2006 Apr;54(4):289-97. doi: 10.1016/j.diagmicrobio.2005.11.002. Epub 2006 Feb 8. Diagn Microbiol Infect Dis. 2006. PMID: 16466900
-
Panfungal PCR and multiplex liquid hybridization for detection of fungi in tissue specimens.J Clin Microbiol. 2000 Nov;38(11):4186-92. doi: 10.1128/JCM.38.11.4186-4192.2000. J Clin Microbiol. 2000. PMID: 11060088 Free PMC article.
-
Rapid identification of yeasts commonly found in positive blood cultures by amplification of the internal transcribed spacer regions 1 and 2.Eur J Clin Microbiol Infect Dis. 2003 Nov;22(11):693-6. doi: 10.1007/s10096-003-1020-5. Epub 2003 Oct 14. Eur J Clin Microbiol Infect Dis. 2003. PMID: 14557922
-
Molecular diagnosis and epidemiology of fungal infections.Med Mycol. 1998;36 Suppl 1:249-57. Med Mycol. 1998. PMID: 9988514 Review.
-
Current status of nonculture methods for diagnosis of invasive fungal infections.Clin Microbiol Rev. 2002 Jul;15(3):465-84. doi: 10.1128/CMR.15.3.465-484.2002. Clin Microbiol Rev. 2002. PMID: 12097252 Free PMC article. Review.
Cited by
-
Rapid identification of yeasts from positive blood culture bottles by pyrosequencing.Eur J Clin Microbiol Infect Dis. 2011 Jan;30(1):21-4. doi: 10.1007/s10096-010-1045-5. Epub 2010 Aug 28. Eur J Clin Microbiol Infect Dis. 2011. PMID: 20803046
-
A sensitive & specific multiplex PCR assay for simultaneous detection of Bacillus anthracis, Yersinia pestis, Burkholderia pseudomallei & Brucella species.Indian J Med Res. 2013;138(1):111-6. Indian J Med Res. 2013. PMID: 24056564 Free PMC article.
-
RELATED FACTORS FOR COLONIZATION BY Candida SPECIES IN THE ORAL CAVITY OF HIV-INFECTED INDIVIDUALS.Rev Inst Med Trop Sao Paulo. 2015 Sep-Oct;57(5):413-9. doi: 10.1590/S0036-46652015000500008. Rev Inst Med Trop Sao Paulo. 2015. PMID: 26603229 Free PMC article.
-
Biosensors and Diagnostics for Fungal Detection.J Fungi (Basel). 2020 Dec 8;6(4):349. doi: 10.3390/jof6040349. J Fungi (Basel). 2020. PMID: 33302535 Free PMC article. Review.
-
Characterization and virulence of yeasts associated with neonatal thrush.BMC Microbiol. 2025 Jun 26;25(1):366. doi: 10.1186/s12866-025-04075-4. BMC Microbiol. 2025. PMID: 40571928 Free PMC article.
References
-
- Ajello, L., and R. J. Hay (ed.). 1998. Medical mycology, p. 1-711. Edward Arnold, London, United Kingdom.
-
- Barnes, R. A., D. W. Denning, E. G. V. Evans, R. J. Hay, C. C. Kibbler, A. G. Prentice, M. D. Richardson, M. M. Roberts, T. R. Rogers, D. C. Speller, D. W. Warnock, and R. E. Warren. 1996. Fungal infections: a survey of laboratory services for diagnosis and treatment. Commun. Dis. Rep. Rev. 6:R69-R75. - PubMed
-
- Buchman, T. G., M. Rossier, W. G. Merz, and P. Charache. 1990. Detection of surgical pathogens by in vitro DNA amplification. Part I. Rapid identification of Candida albicans by in vitro amplification of a fungus-specific gene. Surgery 108:338-347. - PubMed
-
- Burgener-Kairuz, P., J.-P. Zuber, P. Jaunin, T. G. Buchman, J. L. Bille, and M. Rossier. 1994. Rapid detection and identification of Candida albicans and Torulopsis (Candida) glabrata in clinical specimens by species-specific nested PCR amplification of a cytochrome P-450 lanosterol-α-demethylase (L1A1) gene fragment. J. Clin. Microbiol. 32:1902-1907. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

