C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development
- PMID: 12149434
- PMCID: PMC125069
- DOI: 10.1073/pnas.112276199
C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development
Abstract
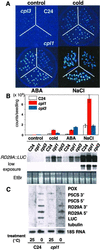
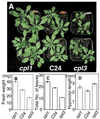
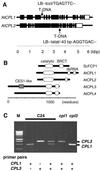
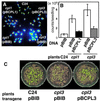
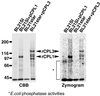
Cold, hyperosmolarity, and abscisic acid (ABA) signaling induce RD29A expression, which is an indicator of the plant stress adaptation response. Two nonallelic Arabidopsis thaliana (ecotype C24) T-DNA insertional mutations, cpl1 and cpl3, were identified based on hyperinduction of RD29A expression that was monitored by using the luciferase (LUC) reporter gene (RD29ALUC) imaging system. Genetic linkage analysis and complementation data established that the recessive cpl1 and cpl3 mutations are caused by T-DNA insertions in AtCPL1 (Arabidopsis C-terminal domain phosphatase-like) and AtCPL3, respectively. Gel assays using recombinant AtCPL1 and AtCPL3 detected innate phosphatase activity like other members of the phylogenetically conserved family that dephosphorylate the C-terminal domain of RNA polymerase II (RNAP II). cpl1 mutation causes RD29ALUC hyperexpression and transcript accumulation in response to cold, ABA, and NaCl treatments, whereas the cpl3 mutation mediates hyperresponsiveness only to ABA. Northern analysis confirmed that LUC transcript accumulation also occurs in response to these stimuli. cpl1 plants accumulate biomass more rapidly and exhibit delayed flowering relative to wild type whereas cpl3 plants grow more slowly and flower earlier than wild-type plants. Hence AtCPL1 and AtCPL3 are negative regulators of stress responsive gene transcription and modulators of growth and development. These results suggest that C-terminal domain phosphatase regulation of RNAP II phosphorylation status is a focal control point of complex processes like plant stress responses and development. AtCPL family members apparently have both unique and overlapping transcriptional regulatory functions that differentiate the signal output that determines the plant response.
Figures
References
-
- Hasegawa P. M., Bressan, R. A., Zhu, J.-K. & Bohnert, H. J. (2000) Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 463-499. - PubMed
-
- Yamaguchi-Shinozaki K., Koizumi, K., Urao, S. & Shinozaki, K. (1992) Plant Cell Physiol. 33 217-224.
-
- Choi H., Hong, J., Ha, J., Kang, J. & Kim, S. Y. (1999) J. Biol. Chem. 275 1723-1730. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases


