Chimeric human CstF-77/Drosophila Suppressor of forked proteins rescue suppressor of forked mutant lethality and mRNA 3' end processing in Drosophila
- PMID: 12149458
- PMCID: PMC124984
- DOI: 10.1073/pnas.162191899
Chimeric human CstF-77/Drosophila Suppressor of forked proteins rescue suppressor of forked mutant lethality and mRNA 3' end processing in Drosophila
Abstract
The Suppressor of forked [Su(f)] protein is the Drosophila homologue of CstF-77, a subunit of human cleavage stimulation factor (CstF) that is required for the first step of the mRNA 3' end processing reaction in vitro. We have addressed directly the role of su(f) in the mRNA 3' end processing reaction in vivo. We show that su(f) is required for the cleavage of pre-mRNA during mRNA 3' end formation. Analysis of the functional complementation between Su(f) and CstF-77 shows that most of the Drosophila protein (85%) can be exchanged for the human protein to produce chimeric CstF-77/Su(f) proteins that rescue lethality and cleavage defect during mRNA 3' end formation in su(f) mutants. Interestingly, we show that a domain in human CstF-77 is limiting for the rescue and that this domain is not able to reproduce protein interactions with the CstF subunits of Drosophila. We also show that chimeric CstF-77/Su(f) proteins that rescue lethality of su(f) mutants cannot restore utilization of a regulated poly(A) site in Drosophila. Taken together, these results demonstrate that CstF-77 and Su(f) have the same function in mRNA 3' end formation in vivo, but that these two proteins are not interchangeable for regulation of poly(A) site utilization.
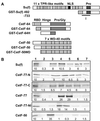
Figures



References
-
- Wahle E. & Rüegsegger, U. (1999) FEMS Microbiol. Rev. 23, 277-295. - PubMed
-
- Bienroth S., Wahle, E., Suter-Crazzolara, C. & Keller, W. (1991) J. Biol. Chem. 266, 19768-19776. - PubMed
-
- Takagaki Y., Manley, J. L., MacDonald, C. C., Wilusz, J. & Shenk, T. (1990) Genes Dev. 4, 2112-2120. - PubMed
-
- Rüegsegger U., Blank, D. & Keller, W. (1998) Mol. Cell 1, 243-253. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

