Global analysis of stress-regulated mRNA turnover by using cDNA arrays
- PMID: 12149460
- PMCID: PMC124989
- DOI: 10.1073/pnas.162212399
Global analysis of stress-regulated mRNA turnover by using cDNA arrays
Abstract
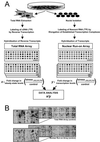
cDNA array technology has proven to be a powerful way to monitor global changes in gene expression patterns. Here, we present an approach that extends the current utility of cDNA arrays to allow the evaluation of the relative roles of transcription and mRNA turnover in governing gene expression on a global basis, compared with current individual gene-by-gene analyses. This method, which involves comparison of large-scale hybridization patterns generated with steady-state mRNA versus newly transcribed (nuclear run-on) RNA, was used to demonstrate the importance of mRNA turnover in regulating gene expression following several conditions of stress.
Figures



Comment in
-
Life after transcription--revisiting the fate of messenger RNA.Trends Genet. 2003 Mar;19(3):113-5. doi: 10.1016/S0168-9525(02)00047-1. Trends Genet. 2003. PMID: 12615000
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

