Competition between histone H1 and HMGN proteins for chromatin binding sites
- PMID: 12151335
- PMCID: PMC1084210
- DOI: 10.1093/embo-reports/kvf156
Competition between histone H1 and HMGN proteins for chromatin binding sites
Abstract
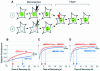
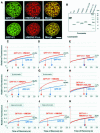
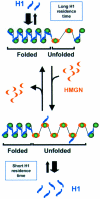
The ability of regulatory factors to access their nucleosomal targets is modulated by nuclear proteins such as histone H1 and HMGN (previously named HMG-14/-17 family) that bind to nucleosomes and either stabilize or destabilize the higher-order chromatin structure. We tested whether HMGN proteins affect the interaction of histone H1 with chromatin. Using microinjection into living cells expressing H1-GFP and photobleaching techniques, we found that wild-type HMGN, but not HMGN point mutants that do not bind to nucleosomes, inhibits the binding of H1 to nucleosomes. HMGN proteins compete with H1 for nucleosome sites but do not displace statically bound H1 from chromatin. Our results provide evidence for in vivo competition among chromosomal proteins for binding sites on chromatin and suggest that the local structure of the chromatin fiber is modulated by a dynamic interplay between nucleosomal binding proteins.
Figures

References
-
- Alfonso P.J., Crippa, M.P., Hayes, J.J. and Bustin, M. (1994) The footprint of chromosomal proteins HMG-14 and HMG-17 on chromatin subunits. J. Mol. Biol., 236, 189–198. - PubMed
-
- Bustin M. (1973) Arrangement of histones in chromatin. Nat. New Biol., 245, 207–209. - PubMed
-
- Bustin M. (2001a) Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci., 26, 152–153. - PubMed
-
- Bustin M. (2001b) Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem. Sci., 26, 431–437. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

