Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcgamma receptor homologs
- PMID: 12163579
- PMCID: PMC136976
- DOI: 10.1128/jvi.76.17.8596-8608.2002
Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcgamma receptor homologs
Abstract
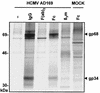
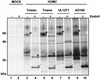
Cellular receptors for the Fc domain of immunoglobulin G (IgG) (FcgammaRs) comprise a family of surface receptors on immune cells connecting humoral and cellular immune responses. Several herpesviruses induce FcgammaR activities in infected cells. Here we identify two distinct human cytomegalovirus (HCMV)-encoded vFcgammaR glycoproteins of 34 and 68 kDa. A panel of HCMV strains exhibited a slight molecular microheterogeneity between Fcgamma-binding proteins, suggesting their viral origin. To locate the responsible genes within the HCMV genome, a large set of targeted HCMV deletion mutants was constructed. The mutant analysis allowed the identification of a spliced UL119-UL118 mRNA to encode vFcgammaR gp68 and TRL11/IRL11 to encode vFcgammaR gp34. Both vFcgammaRs are surface resident type I transmembrane glycoproteins. Significant relatedness of sequences in the extracellular chain of gpUL119-118 and gpTRL11 with particular immunoglobulin supergene family domains present in FcgammaR I and FcgammaRs II/III, respectively, indicates a different ancestry and function of gpUL119-118 and gpTRL11. The HCMV-encoded vFcgammaRs highlight an impressive diversification and redundancy of FcgammaR structures.
Figures

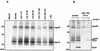








References
-
- Amigorena, S., and C. Bonnerot. 1999. Fc receptor signaling and trafficking: a connection for antigen processing. Immunol. Rev. 172:279-284. - PubMed
-
- Antonsson, A., and P. J. Johansson. 2001. Binding of human and animal immunoglobulins to the IgG Fc receptor induced by human cytomegalovirus. J. Gen. Virol. 82:1137-1145. - PubMed
-
- Burmeister, W. P., L. N. Gastinel, N. E. Simister, M. L. Blum, and P. J. Bjorkman. 1994. Crystal structure at 2.2 A resolution of the MHC-related neonatal Fc receptor. Nature 372:336-343. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

