Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm
- PMID: 12163602
- PMCID: PMC136978
- DOI: 10.1128/jvi.76.17.8820-8833.2002
Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm
Abstract
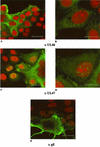
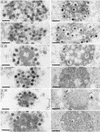
Proteins encoded by the UL46 and UL47 genes of herpes simplex virus type 1 (HSV-1) constitute major components of the viral tegument. However, their functions have so far not been elucidated in detail. By use of monospecific antisera directed against bacterially expressed glutathione-S-transferase fusion proteins, the homologous UL46 and UL47 proteins of the alphaherpesvirus pseudorabies virus (PrV) were identified in virus-infected cells and in virions. The PrV UL46 gene product of 693 amino acids (aa) exhibits an apparent molecular mass of 95 kDa, whereas the UL47 product of 750 aa was identified as a 97-kDa protein. Both are present in purified virions, correlating with their role as tegument proteins. Immunofluorescence analysis by confocal laser scan microscopy showed that late in infection the UL46 product is detectable in the cytoplasm, whereas the UL47 product was observed to be diffuse in the cytoplasm and speckled in the nucleus. Virus mutants lacking either the UL46 or the UL47 gene or both were isolated on noncomplementing cells, demonstrating that these genes either singly or in combination are not required for productive viral replication. However, plaque sizes were decreased. Interestingly, in one-step growth analysis, UL47 deletion mutants exhibited an approximately 10-fold decrease in final titers, whereas the UL46 deletion mutant was not affected. This finding correlated with ultrastructural observations which showed unimpaired virion morphogenesis in the absence of the UL46 protein, whereas in the absence of the UL47 protein intracytoplasmic aggregates of partially tegumented capsids were observed. In summary, we identified the PrV UL46 and UL47 proteins and show that the UL47 protein plays an important role in virion assembly in the cytoplasm.
Figures






References
-
- Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. F. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature (London) 310:207-211. - PubMed
-
- Barker, D., and B. Roizman. 1990. Identification of three genes nonessential for growth in cell culture near the right terminus of the unique sequences of long component of herpes simplex virus 1. Virology 177:684-692. - PubMed
-
- Blaho, J. A., C. Mitchell, and B. Roizman. 1994. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47 and UL49 gene products. J. Biol. Chem. 269:17401-17410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

