Molecular analysis of three Ljungan virus isolates reveals a new, close-to-root lineage of the Picornaviridae with a cluster of two unrelated 2A proteins
- PMID: 12163611
- PMCID: PMC137002
- DOI: 10.1128/jvi.76.17.8920-8930.2002
Molecular analysis of three Ljungan virus isolates reveals a new, close-to-root lineage of the Picornaviridae with a cluster of two unrelated 2A proteins
Abstract
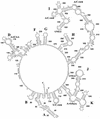
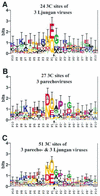
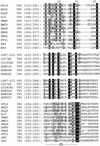
Ljungan virus (LV) is a suspected human pathogen recently isolated from bank voles (Clethrionomys glareolus). In the present study, it is revealed through comparative sequence analysis that three newly determined Swedish LV genomes are closely related and possess a deviant picornavirus-like organization: 5' untranslated region-VP0-VP3-VP1-2A1-2A2-2B-2C-3A-3B-3C-3D-3' untranslated region. The LV genomes and the polyproteins encoded by them exhibit several exceptional features, such as the absence of a predicted maturation cleavage of VP0, a conserved sequence determinant in VP0 that is typically found in VP1 of other picornaviruses, and a cluster of two unrelated 2A proteins. The 2A1 protein is related to the 2A protein of cardio-, erbo-, tescho-, and aphthoviruses, and the 2A2 protein is related to the 2A protein of parechoviruses, kobuviruses, and avian encephalomyelitis virus. The unprecedented association of two structurally different 2A proteins is a feature never previously observed among picornaviruses and implies that their functions are not mutually exclusive. Secondary polyprotein processing of the LV polyprotein is mediated by proteinase 3C (3C(pro)) possessing canonical affinity to Glu and Gln at the P1 position and small amino acid residues at the P1' position. In addition, LV 3C(pro) appears to have unique substrate specificity to Asn, Gln, and Asp and to bulky hydrophobic residues at the P2 and P4 positions, respectively. Phylogenetic analysis suggests that LVs form a separate division, which, together with the Parechovirus genus, has branched off the picornavirus tree most closely to its root. The presence of two 2A proteins indicates that some contemporary picornaviruses with a single 2A may have evolved from the ancestral multi-2A picornavirus.
Figures



References
-
- Auvinen, P., and T. Hyypiä. 1990. Echoviruses include genetically distinct serotypes. J. Gen. Virol. 71:2133-2139. - PubMed
-
- Bertioli, D. 1997. Rapid amplification of cDNA ends, p. 233-238. In B. A. White (ed.), PCR cloning protocols: from molecular cloning to genetic engineering, vol. 67. Humana Press Inc., Totowa, N.J.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

