tRNA-mediated transcription antitermination in vitro: codon-anticodon pairing independent of the ribosome
- PMID: 12165569
- PMCID: PMC123220
- DOI: 10.1073/pnas.162366799
tRNA-mediated transcription antitermination in vitro: codon-anticodon pairing independent of the ribosome
Abstract
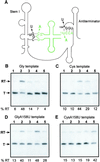
Uncharged tRNA acts as the effector for transcription antitermination of genes in the T box family in Bacillus subtilis and other Gram-positive bacteria. Genetic studies suggested that expression of these genes is induced by stabilization of an antiterminator element in the leader RNA of each target gene by the cognate uncharged tRNA. The specificity of the tRNA response is dependent on a single codon in the leader, which was postulated to pair with the anticodon of the corresponding tRNA. It was not known whether the leader RNA-tRNA interaction requires additional factors. We show here that tRNA-dependent antitermination occurs in vitro in a purified transcription system, in the absence of ribosomes or accessory factors, demonstrating that the RNA-RNA interaction is sufficient to control gene expression by antitermination. The tRNA response exhibits similar specificity in vivo and in vitro, and the antitermination reaction in vitro is independent of NusA and functions with either B. subtilis or Escherichia coli RNA polymerase.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

