Biogenesis of yeast telomerase depends on the importin mtr10
- PMID: 12167699
- PMCID: PMC134008
- DOI: 10.1128/MCB.22.17.6046-6055.2002
Biogenesis of yeast telomerase depends on the importin mtr10
Abstract
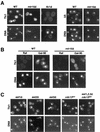
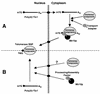
Telomerase is a ribonucleoprotein particle (RNP) involved in chromosome end replication, but its biogenesis is poorly understood. The RNA component of yeast telomerase (Tlc1) is synthesized as a polyadenylated precursor and then processed to a mature poly(A)- form. We report here that the karyopherin Mtr10p is required for the normal accumulation of mature Tlc1 and its proper localization to the nucleus. Neither TLC1 transcription nor the stability of poly(A)- Tlc1 is significantly affected in mtr10delta cells. Tlc1 was mostly nuclear in a wild-type background, and this localization was not affected by mutations in other telomerase components. Strikingly, in the absence of Mtr10p, Tlc1 was found dispersed throughout the entire cell. Our results are compatible with two alternative models. First, Mtr10p may import a cytoplasmic complex containing Tlc1 and perhaps other components of telomerase, and shuttling of Tlc1 from the nucleus to the cytoplasm and back may be necessary for the biogenesis of telomerase (the "shuttling" model). Second, Mtr10p may be necessary for the nuclear import of some enzyme needed for the nuclear processing and maturation of Tlc1, and in the absence of this maturation, poly(A)+ Tlc1 is aberrantly exported to the cytoplasm (the "processing enzyme" model).
Figures


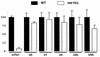


References
-
- Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. - PubMed
-
- Brown, T. 1994. Preparation and analysis of DNA, p. 2.9.7-2.9.8. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. More, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
-
- Collart, M. A., and S. Oliviero. 1994. Saccharomyces cerevisiae, p. 13.12.1-13.12.2. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. More, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
