Hypophosphorylation of Mdm2 augments p53 stability
- PMID: 12167711
- PMCID: PMC134018
- DOI: 10.1128/MCB.22.17.6170-6182.2002
Hypophosphorylation of Mdm2 augments p53 stability
Abstract
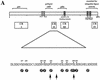
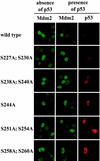
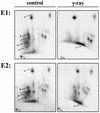
The Mdm2 protein mediates ubiquitylation and degradation of p53 and is a key regulator of this tumor suppressor. More recently, it has been shown that Mdm2 is highly phosphorylated within its central acidic domain. In order to address the issue of how these modifications might regulate Mdm2 function, putative phosphorylation sites within this domain were substituted, individually or in pairs, with alanine residues. Mutants with serine-to-alanine substitutions between residues 244 and 260 abolished or at least reduced the capacity of Mdm2 to promote p53 degradation. In each case, loss of degradation function was independent of the ability to bind to p53 or p14ARF. Moreover, each of the Mdm2 mutants completely retained the capacity to act as a ubiquitin ligase in vivo. Thus, ubiquitylation and degradation can be uncoupled. Two-dimensional phosphopeptide mapping coupled with the use of phospho-specific antibodies revealed that Mdm2 is phosphorylated physiologically at several sites within this region, consistent with the idea that phosphorylation is important for Mdm2 activity. Strikingly, treatment of cells with ionizing radiation resulted in a significant decrease in the phosphorylation of residues that are important for p53 turnover. This hypophosphorylation preceded p53 accumulation. These findings indicate that Mdm2 contributes an additional function toward the degradation of p53 that is distinct from its ubiquitin ligase activity and is regulated by phosphorylation. Our model suggests that hypophosphorylation of Mdm2 in response to ionizing irradiation inactivates this novel function, thereby contributing to p53 stabilization.
Figures



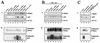




References
-
- Argentini, M., N. Barboule, and B. Wasylyk. 2001. The contribution of the acidic domain of MDM2 to p53 and MDM2 stability. Oncogene 20:1267-1275. - PubMed
-
- Barak, Y., E. Gottlieb, G. T. Juven, and M. Oren. 1994. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with non-identical translation potential. Genes Dev. 8:1739-1749. - PubMed
-
- Bottger, A., V. Bottger, A. Sparks, W.-L. Liu, S. F. Howard, and D. P. Lane. 1997. Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr. Biol. 7:860-869. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
