C-terminal regions of the human telomerase catalytic subunit essential for in vivo enzyme activity
- PMID: 12167716
- PMCID: PMC134020
- DOI: 10.1128/MCB.22.17.6234-6246.2002
C-terminal regions of the human telomerase catalytic subunit essential for in vivo enzyme activity
Abstract
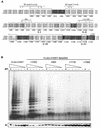
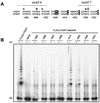
Most human cancer cells are thought to acquire the ability to divide beyond the capacity of normal somatic cells through illegitimately activating the gene hTERT, which encodes the catalytic subunit of telomerase. While telomerase reverse transcriptase (TERT) is conserved in most eukaryotes, mounting evidence suggests that the C terminus of the human protein may have functions unique to higher eukaryotes. To search for domains responsible for such functions, we assayed a panel of tandem substitution mutations encompassing this region of human TERT for in vitro and in vivo functionality. We found four clusters of mutations that inactivated the biochemical and biological functions of telomerase, separated by mutations that had little or no effect on enzyme activity. We also identified a region where mutations generate catalytically active but biologically inert proteins. This C-terminal region that dissociates activities of telomerase (C-DAT) does not appear to be involved in nuclear localization or protein multimerization. Instead, it appears that the C-DAT region is involved in a step of in vivo telomere synthesis after the assembly of a catalytically active enzyme. Intriguingly, all of the described regions reside in a portion of TERT that is dispensable for cellular viability in yeast, arguing for a divergent role of the C terminus in higher eukaryotes.
Figures
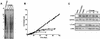
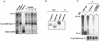

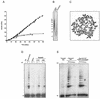

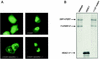
References
-
- Andersson, S., D. L. Davis, H. Dahlback, H. Jornvall, and D. W. Russell. 1989. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J. Biol. Chem. 264:8222-8229. - PubMed
-
- Arai, K., K. Masutomi, S. Khurts, S. Kaneko, K. Kobayashi, and S. Murakami. 2002. Two independent regions of human telomerase reverse transcriptase are important for its oligomerization and telomerase activity. J. Biol. Chem. 277:8538-8544. - PubMed
-
- Bachand, F., and C. Autexier. 1999. Functional reconstitution of human telomerase expressed in Saccharomyces cerevisiae. J. Biol. Chem. 274:38027-38031. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
