mRNA decay in Escherichia coli comes of age
- PMID: 12169588
- PMCID: PMC135271
- DOI: 10.1128/JB.184.17.4658-4665.2002
mRNA decay in Escherichia coli comes of age
Figures
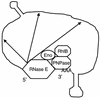
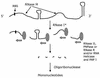

References
-
- Achord, D., and D. Kennell. 1974. Metabolism of messenger RNA from the gal operon of Escherichia coli. J. Mol. Biol. 90:581-599. - PubMed
-
- Apirion, D. 1973. Degradation of RNA in Escherichia coli: a hypothesis. Mol. Gen. Genet. 122:313-322. - PubMed
-
- August, J., P. J. Ortiz, and J. Hurwitz. 1962. Ribonucleic acid-dependent ribonucleotide incorporation. I. Purification and properties of the enzyme. J. Biol. Chem. 237:3786-3793. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

