Transposase-dependent formation of circular IS256 derivatives in Staphylococcus epidermidis and Staphylococcus aureus
- PMID: 12169594
- PMCID: PMC135277
- DOI: 10.1128/JB.184.17.4709-4714.2002
Transposase-dependent formation of circular IS256 derivatives in Staphylococcus epidermidis and Staphylococcus aureus
Abstract
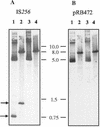
IS256 is a highly active insertion sequence (IS) element of multiresistant staphylococci and enterococci. Here we show that, in a Staphylococcus epidermidis clinical isolate, as well as in recombinant Staphylococcus aureus and Escherichia coli carrying a single IS256 insertion on a plasmid, IS256 excises as an extrachromosomal circular DNA molecule. First, circles were identified that contained a complete copy of IS256. In this case, the sequence connecting the left and right ends of IS256 was derived from flanking DNA sequences of the parental genetic locus. Second, circle junctions were detected in which one end of IS256 was truncated. Nucleotide sequencing of circle junctions revealed that (i) either end of IS256 can attack the opposite terminus and (ii) the circle junctions vary significantly in size. Upon deletion of the IS256 open reading frame at the 3' end and site-directed mutageneses of the putative DDE motif, circular IS256 molecules were no longer detectable, which implicates the IS256-encoded transposase protein with the circularization of the element.
Figures

Similar articles
-
Characterization of the transposase encoded by IS256, the prototype of a major family of bacterial insertion sequence elements.J Bacteriol. 2010 Aug;192(16):4153-63. doi: 10.1128/JB.00226-10. Epub 2010 Jun 11. J Bacteriol. 2010. PMID: 20543074 Free PMC article.
-
A transposase-independent mechanism gives rise to precise excision of IS256 from insertion sites in Staphylococcus epidermidis.J Bacteriol. 2008 Feb;190(4):1488-90. doi: 10.1128/JB.01290-07. Epub 2007 Dec 7. J Bacteriol. 2008. PMID: 18065530 Free PMC article.
-
Typing of Staphylococcus aureus and Staphylococcus epidermidis strains by PCR analysis of inter-IS256 spacer length polymorphisms.J Clin Microbiol. 1997 Oct;35(10):2580-7. doi: 10.1128/jcm.35.10.2580-2587.1997. J Clin Microbiol. 1997. PMID: 9316911 Free PMC article.
-
Success through diversity - how Staphylococcus epidermidis establishes as a nosocomial pathogen.Int J Med Microbiol. 2010 Aug;300(6):380-6. doi: 10.1016/j.ijmm.2010.04.011. Epub 2010 May 6. Int J Med Microbiol. 2010. PMID: 20451447 Review.
-
Genetic manipulation of Staphylococci-breaking through the barrier.Front Cell Infect Microbiol. 2012 Apr 12;2:49. doi: 10.3389/fcimb.2012.00049. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22919640 Free PMC article. Review.
Cited by
-
Phase and antigenic variation in bacteria.Clin Microbiol Rev. 2004 Jul;17(3):581-611, table of contents. doi: 10.1128/CMR.17.3.581-611.2004. Clin Microbiol Rev. 2004. PMID: 15258095 Free PMC article. Review.
-
Chromosomal context directs high-frequency precise excision of IS492 in Pseudoalteromonas atlantica.Proc Natl Acad Sci U S A. 2007 Feb 6;104(6):1901-6. doi: 10.1073/pnas.0608633104. Epub 2007 Jan 30. Proc Natl Acad Sci U S A. 2007. PMID: 17264213 Free PMC article.
-
The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides.Infect Immun. 2004 Feb;72(2):1210-5. doi: 10.1128/IAI.72.2.1210-1215.2004. Infect Immun. 2004. PMID: 14742578 Free PMC article.
-
Targeted IS-element sequencing uncovers transposition dynamics during selective pressure in enterococci.PLoS Pathog. 2023 Jun 2;19(6):e1011424. doi: 10.1371/journal.ppat.1011424. eCollection 2023 Jun. PLoS Pathog. 2023. PMID: 37267422 Free PMC article.
-
Structures of ISCth4 transpososomes reveal the role of asymmetry in copy-out/paste-in DNA transposition.EMBO J. 2021 Jan 4;40(1):e105666. doi: 10.15252/embj.2020105666. Epub 2020 Oct 2. EMBO J. 2021. PMID: 33006208 Free PMC article.
References
-
- Biery, M. C., M. Lopata, and N. L. Craig. 2000. A minimal system for Tn7 transposition: the transposon-encoded proteins TnsA and TnsB can execute DNA breakage and joining reactions that generate circularized Tn7 species. J. Mol. Biol. 297:25-37. - PubMed
-
- Byrne, M. E., D. A. Rouch, and R. A. Skurray. 1989. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene 81:361-367. - PubMed
-
- Haren, L., B. Ton-Hoang, and M. Chandler. 1999. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 53:245-281. - PubMed
-
- Kiss, J., and F. Olasz. 1999. Formation and transposition of the covalently closed IS30 circle: the relation between tandem dimers and monomeric circles. Mol. Microbiol. 34:37-52. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

