Site-specific recombination system encoded by toluene catabolic transposon Tn4651
- PMID: 12169600
- PMCID: PMC135285
- DOI: 10.1128/JB.184.17.4757-4766.2002
Site-specific recombination system encoded by toluene catabolic transposon Tn4651
Abstract
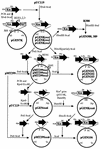
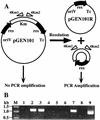
The 56-kb class II toluene catabolic transposon Tn4651 from Pseudomonas putida plasmid pWW0 is unique in that (i) its efficient resolution requires, in addition to the 0.2-kb resolution (res) site, the two gene products TnpS and TnpT and (ii) the 2.4-kb tnpT-res-tnpS region is 48 kb apart from the tnpA gene (M. Tsuda, K.-I. Minegishi, and T. Iino, J. Bacteriol. 171:1386-1393, 1989). Detailed analysis of the 2.4-kb region revealed that the tnpS and tnpT genes encoding the putative 323- and 332-amino-acid proteins, respectively, were transcribed divergently with an overlapping 59-bp sequence in the 203-bp res site. The motifs (the R-H-R-Y tetrad in domains I and II with proper spacing) commonly conserved in the integrase family of site-specific recombinases were found in TnpS. In contrast, TnpT did not show any significant amino acid sequence homology to the other proteins that are directly or indirectly involved in recombination. Analysis of site-specific recombination under the Escherichia coli recA cells indicated that (i) the site-specific resolution between the two copies of the res site on a single molecule was catalyzed by TnpS, (ii) the functional res site was located within a 95-bp segment, and (iii) TnpT appeared to have the role of enhancing the site-specific resolution. It was also found that TnpS catalyzed the site-specific recombination between the res sites located at two different molecules to form a cointegrate molecule. Site-specific mutagenesis of the conserved tyrosine residue in TnpS led to the loss of both the resolution and the integration activities, indicating that such a residue took part in both types of recombination.
Figures


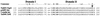
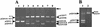
Similar articles
-
Cointegrate-resolution of toluene-catabolic transposon Tn4651: determination of crossover site and the segment required for full resolution activity.Plasmid. 2013 Jan;69(1):24-35. doi: 10.1016/j.plasmid.2012.07.004. Epub 2012 Aug 1. Plasmid. 2013. PMID: 22878084
-
Toluene transposons Tn4651 and Tn4653 are class II transposons.J Bacteriol. 1989 Mar;171(3):1386-93. doi: 10.1128/jb.171.3.1386-1393.1989. J Bacteriol. 1989. PMID: 2537816 Free PMC article.
-
Genetic analysis of a transposon carrying toluene degrading genes on a TOL plasmid pWW0.Mol Gen Genet. 1987 Dec;210(2):270-6. doi: 10.1007/BF00325693. Mol Gen Genet. 1987. PMID: 2830457
-
Structure and function of the shufflon in plasmid R64.Adv Biophys. 2004;38:183-213. Adv Biophys. 2004. PMID: 15493334 Review.
-
Conjugative transposition.Annu Rev Microbiol. 1995;49:367-97. doi: 10.1146/annurev.mi.49.100195.002055. Annu Rev Microbiol. 1995. PMID: 8561465 Review.
Cited by
-
Transcriptome analysis of Pseudomonas putida KT2440 harboring the completely sequenced IncP-7 plasmid pCAR1.J Bacteriol. 2007 Oct;189(19):6849-60. doi: 10.1128/JB.00684-07. Epub 2007 Aug 3. J Bacteriol. 2007. PMID: 17675379 Free PMC article.
-
Characterization of Halomonas sp. ZM3 isolated from the Zelazny Most post-flotation waste reservoir, with a special focus on its mobile DNA.BMC Microbiol. 2013 Mar 14;13:59. doi: 10.1186/1471-2180-13-59. BMC Microbiol. 2013. PMID: 23497212 Free PMC article.
-
Genomic and functional analysis of the IncP-9 naphthalene-catabolic plasmid NAH7 and its transposon Tn4655 suggests catabolic gene spread by a tyrosine recombinase.J Bacteriol. 2006 Jun;188(11):4057-67. doi: 10.1128/JB.00185-06. J Bacteriol. 2006. PMID: 16707697 Free PMC article.
-
Annotation and Comparative Genomics of Prokaryotic Transposable Elements.Methods Mol Biol. 2024;2802:189-213. doi: 10.1007/978-1-0716-3838-5_8. Methods Mol Biol. 2024. PMID: 38819561
-
Towards a more accurate annotation of tyrosine-based site-specific recombinases in bacterial genomes.Mob DNA. 2012 Apr 13;3(1):6. doi: 10.1186/1759-8753-3-6. Mob DNA. 2012. PMID: 22502997 Free PMC article.
References
-
- Amann, E., J. Brosius, and M. Ptashne. 1983. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene 25:167-178. - PubMed
-
- Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused protein in Escherichia coli. Gene 69:301-315. - PubMed
-
- Assinder, S. J., and P. A. Williams. 1990. The TOL plasmids: determinants of the catabolism of toluene and the xylenes. Adv. Microb. Physiol. 31:1-69. - PubMed
-
- Avila, P., and F. de la Cruz. 1988. Physical and genetic mapping of the IncW plasmid R388. Plasmid 20:155-157. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

