Effects of the P1 plasmid centromere on expression of P1 partition genes
- PMID: 12169611
- PMCID: PMC135286
- DOI: 10.1128/JB.184.17.4857-4867.2002
Effects of the P1 plasmid centromere on expression of P1 partition genes
Abstract
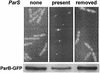
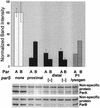
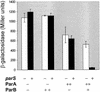
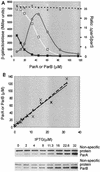
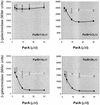
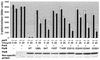
The partition operon of P1 plasmid encodes two proteins, ParA and ParB, required for the faithful segregation of plasmid copies to daughter cells. The operon is followed by a centromere analog, parS, at which ParB binds. ParA, a weak ATPase, represses the par promoter most effectively in its ADP-bound form. ParB can recruit ParA to parS, stimulate its ATPase, and significantly stimulate the repression. We report here that parS also participates in the regulation of expression of the par genes. A single chromosomal parS was shown to augment repression of several copies of the par promoter by severalfold. The repression increase was sensitive to the levels of ParA and ParB and to their ratio. The increase may be attributable to a conformational change in ParA mediated by the parS-ParB complex, possibly acting catalytically. We also observed an in cis effect of parS which enhanced expression of parB, presumably due to a selective modulation of the mRNA level. Although ParB had been earlier found to spread into and silence genes flanking parS, silencing of the par operon by ParB spreading was not significant. Based upon analogies between partitioning and septum placement, we speculate that the regulatory switch controlled by the parS-ParB complex might be essential for partitioning itself.
Figures

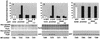
Similar articles
-
Molecular analysis of the pRA2 partitioning region: ParB autoregulates parAB transcription and forms a nucleoprotein complex with the plasmid partition site, parS.Mol Microbiol. 2001 May;40(3):621-33. doi: 10.1046/j.1365-2958.2001.02405.x. Mol Microbiol. 2001. PMID: 11359568
-
P1 ParA interacts with the P1 partition complex at parS and an ATP-ADP switch controls ParA activities.EMBO J. 1999 Mar 1;18(5):1415-24. doi: 10.1093/emboj/18.5.1415. EMBO J. 1999. PMID: 10064607 Free PMC article.
-
Transcriptional interference by a complex formed at the centromere-like partition site of plasmid P1.J Bacteriol. 2002 May;184(9):2447-54. doi: 10.1128/JB.184.9.2447-2454.2002. J Bacteriol. 2002. PMID: 11948158 Free PMC article.
-
The bacterial ParA-ParB partitioning proteins.J Biotechnol. 2001 Sep 13;91(1):1-34. doi: 10.1016/s0168-1656(01)00293-0. J Biotechnol. 2001. PMID: 11522360 Review.
-
Prokaryotic ParA-ParB-parS system links bacterial chromosome segregation with the cell cycle.Plasmid. 2012 Jan;67(1):1-14. doi: 10.1016/j.plasmid.2011.08.003. Epub 2011 Sep 6. Plasmid. 2012. PMID: 21924286 Review.
Cited by
-
KorB switching from DNA-sliding clamp to repressor mediates long-range gene silencing in a multi-drug resistance plasmid.Nat Microbiol. 2025 Feb;10(2):448-467. doi: 10.1038/s41564-024-01915-3. Epub 2025 Jan 23. Nat Microbiol. 2025. PMID: 39849085 Free PMC article.
-
Centromere anatomy in the multidrug-resistant pathogen Enterococcus faecium.Proc Natl Acad Sci U S A. 2008 Feb 12;105(6):2151-6. doi: 10.1073/pnas.0704681105. Epub 2008 Feb 1. Proc Natl Acad Sci U S A. 2008. PMID: 18245388 Free PMC article.
-
Addressing the role of centromere sites in activation of ParB proteins for partition complex assembly.PLoS One. 2020 May 7;15(5):e0226472. doi: 10.1371/journal.pone.0226472. eCollection 2020. PLoS One. 2020. PMID: 32379828 Free PMC article.
-
Catching a Walker in the Act-DNA Partitioning by ParA Family of Proteins.Front Microbiol. 2022 May 26;13:856547. doi: 10.3389/fmicb.2022.856547. eCollection 2022. Front Microbiol. 2022. PMID: 35694299 Free PMC article. Review.
-
Three ParA Dimers Cooperatively Assemble on Type Ia Partition Promoters.Genes (Basel). 2021 Aug 28;12(9):1345. doi: 10.3390/genes12091345. Genes (Basel). 2021. PMID: 34573327 Free PMC article.
References
-
- Austin, S., and K. Nordström. 1990. Partition-mediated incompatibility of bacterial plasmids. Cell 60:351-354. - PubMed
-
- Autret, S., R. Nair, and J. Errington. 2001. Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein. Mol. Microbiol. 41:743-755. - PubMed
-
- Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

