Surface loop motion in FepA
- PMID: 12169616
- PMCID: PMC135268
- DOI: 10.1128/JB.184.17.4906-4911.2002
Surface loop motion in FepA
Abstract
Using a lysine-specific cleavable cross-linking reagent ethylene glycolbis(sulfosuccimidylsuccinate) (Sulfo-EGS), we studied conformational motion in the surface loops of Escherichia coli FepA during its transport of the siderophore ferric enterobactin. Site-directed mutagenesis determined that Sulfo-EGS reacted with two lysines, K332 and K483, and at least two other unidentified Lys residues in the surface loops of the outer membrane protein. The reagent cross-linked K483 in FepA L7 to either K332 in L5, forming a product that we designated band 1, or to the major outer membrane proteins OmpF, OmpC, and OmpA, forming band 2. Ferric enterobactin binding to FepA did not prevent modification of K483 by Sulfo-EGS but blocked its cross-linking to OmpF/C and OmpA and reduced its coupling to K332. These data show that the loops of FepA undergo conformational changes in vivo, with an approximate magnitude of 15 A, from a ligand-free open state to a ligand-bound closed state. The coupling of FepA L7 to OmpF, OmpC, or OmpA was TonB independent and was unaffected by the uncouplers CCCP (carbonyl cyanide m-chlorophenylhydrazone) and DNP (2,4-dinitrophenol) but completely inhibited by cyanide.
Figures
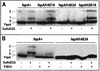

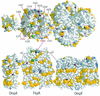
References
-
- Abdella, P. M., P. K. Smith, and G. P. Royer. 1979. A new cleavable reagent for cross-linking and reversible immobilization of proteins. Biochem. Biophys. Res. Commun. 87:734-742. - PubMed
-
- Ames, G. F. 1974. Resolution of bacterial proteins by polyacrylamide gel electrophoresis on slabs: membrane, soluble, and periplasmic fractions. J. Biol. Chem. 249:634-644. - PubMed
-
- Bos, C., D. Lorenzen, and V. Braun. 1998. Specific in vivo labeling of cell surface-exposed protein loops: reactive cysteines in the predicted gating loop mark a ferrichrome binding site and a ligand-induced conformational change of the Escherichia coli FhuA protein. J. Bacteriol. 180:605-613. - PMC - PubMed
-
- Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

