LasR, a transcriptional activator of Pseudomonas aeruginosa virulence genes, functions as a multimer
- PMID: 12169617
- PMCID: PMC135272
- DOI: 10.1128/JB.184.17.4912-4919.2002
LasR, a transcriptional activator of Pseudomonas aeruginosa virulence genes, functions as a multimer
Abstract
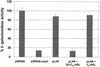
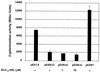
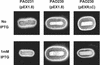
The Pseudomonas aeruginosa LasR protein functions in concert with N-3-oxo-dodecanoyl-L-homoserine lactone (3O-C(12)-HSL) to coordinate the expression of target genes, including many genes that encode virulence factors, with cell density. We used a LexA-based protein interaction assay to demonstrate that LasR forms multimers only when 3O-C(12)-HSL is present. A series of LasR molecules containing internal deletions or substitutions in single, conserved amino acid residues indicated that the N-terminal portion of LasR is required for multimerization. Studies performed with these mutant versions of LasR demonstrated that the ability of LasR to multimerize correlates with its ability to function as a transcriptional activator of lasI, a gene known to be tightly regulated by the LasR-3O-C(12)-HSL regulatory system. A LasR molecule that carries a C-terminal deletion can function as a dominant-negative mutant in P. aeruginosa, as shown by its ability to decrease expression of lasB, another LasR-3O-C(12)-HSL target gene. Taken together, our data strongly support the hypothesis that LasR functions as a multimer in vivo.
Figures
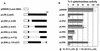

Similar articles
-
Inactivation of the quorum-sensing transcriptional regulators LasR or RhlR does not suppress the expression of virulence factors and the virulence of Pseudomonas aeruginosa PAO1.Microbiology (Reading). 2019 Apr;165(4):425-432. doi: 10.1099/mic.0.000778. Epub 2019 Feb 1. Microbiology (Reading). 2019. PMID: 30707095
-
Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression.J Bacteriol. 2002 May;184(10):2576-86. doi: 10.1128/JB.184.10.2576-2586.2002. J Bacteriol. 2002. PMID: 11976285 Free PMC article.
-
Pseudomonas aeruginosa quorum-sensing response in the absence of functional LasR and LasI proteins: the case of strain 148, a virulent dolphin isolate.FEMS Microbiol Lett. 2017 Jul 3;364(12). doi: 10.1093/femsle/fnx119. FEMS Microbiol Lett. 2017. PMID: 28591849
-
The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS.J Bacteriol. 2000 Nov;182(22):6401-11. doi: 10.1128/JB.182.22.6401-6411.2000. J Bacteriol. 2000. PMID: 11053384 Free PMC article.
-
The effect of specific rhlA-las-box mutations on DNA binding and gene activation by Pseudomonas aeruginosa quorum-sensing transcriptional regulators RhlR and LasR.FEMS Microbiol Lett. 2014 Jul;356(2):217-25. doi: 10.1111/1574-6968.12505. FEMS Microbiol Lett. 2014. PMID: 24935161
Cited by
-
Biological and clinical significance of quorum sensing alkylquinolones: current analytical and bioanalytical methods for their quantification.Anal Bioanal Chem. 2021 Jul;413(18):4599-4618. doi: 10.1007/s00216-021-03356-x. Epub 2021 May 7. Anal Bioanal Chem. 2021. PMID: 33959788 Review.
-
Pseudomonas aeruginosa PA80 is a cystic fibrosis isolate deficient in RhlRI quorum sensing.Sci Rep. 2021 Mar 11;11(1):5729. doi: 10.1038/s41598-021-85100-0. Sci Rep. 2021. PMID: 33707533 Free PMC article.
-
Additive Effects of Quorum Sensing Anti-Activators on Pseudomonas aeruginosa Virulence Traits and Transcriptome.Front Microbiol. 2018 Jan 9;8:2654. doi: 10.3389/fmicb.2017.02654. eCollection 2017. Front Microbiol. 2018. PMID: 29375519 Free PMC article.
-
Novel Insight of Transcription Factor PtrA on Pathogenicity and Carbapenems Resistance in Pseudomonas aeruginosa.Infect Drug Resist. 2022 Aug 4;15:4213-4227. doi: 10.2147/IDR.S371597. eCollection 2022. Infect Drug Resist. 2022. PMID: 35959145 Free PMC article.
-
Cobalt Complex with Thiazole-Based Ligand as New Pseudomonas aeruginosa Quorum Quencher, Biofilm Inhibitor and Virulence Attenuator.Molecules. 2018 Jun 8;23(6):1385. doi: 10.3390/molecules23061385. Molecules. 2018. PMID: 29890626 Free PMC article.
References
-
- Barik, S. 1996. Site-directed mutagenesis in vitro by megaprimer PCR, p. 203-216. In M. K. Trower (ed.), In vitro mutagenesis protocols, vol. 57. Humana Press, Totowa, N.J. - PubMed
-
- Choi, S. H., and E. P. Greenberg. 1992. Genetic evidence for multimerization of LuxR, the transcriptional activator of Vibrio fischeri luminescence. Mol. Mar. Biol. Biotechnol. 1:408-413.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

