Structure of the periplasmic domain of Pseudomonas aeruginosa TolA: evidence for an evolutionary relationship with the TonB transporter protein
- PMID: 12169623
- PMCID: PMC126161
- DOI: 10.1093/emboj/cdf417
Structure of the periplasmic domain of Pseudomonas aeruginosa TolA: evidence for an evolutionary relationship with the TonB transporter protein
Abstract
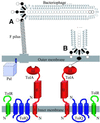
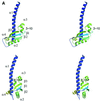
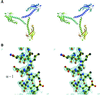
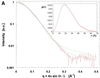
The crystal structure of the C-terminal domain III of Pseudomonas aeruginosa TolA has been determined at 1.9 A resolution. The fold is similar to that of the corresponding domain of Escherichia coli TolA, despite the limited amino acid sequence identity of the two proteins (20%). A pattern was discerned that conserves the fold of domain III within the wider TolA family and, moreover, reveals a relationship between TolA domain III and the C-terminal domain of the TonB transporter proteins. We propose that the TolA and TonB C-terminal domains have a common evolutionary origin and are related by means of domain swapping, with interesting mechanistic implications. We have also determined the overall shape of the didomain, domains II + III, of P.aeruginosa TolA by solution X-ray scattering. The molecule is monomeric-its elongated, stalk shape can accommodate the crystal structure of domain III at one end, and an elongated helical bundle within the portion corresponding to domain II. Based on these data, a model for the periplasmic domains of P.aeruginosa TolA is presented that may explain the inferred allosteric properties of members of the TolA family. The mechanisms of TolA-mediated entry of bateriophages in P.aeruginosa and E.coli are likely to be similar.
Figures









Similar articles
-
Filamentous phage infection: required interactions with the TolA protein.J Bacteriol. 1997 Oct;179(20):6464-71. doi: 10.1128/jb.179.20.6464-6471.1997. J Bacteriol. 1997. PMID: 9335297 Free PMC article.
-
Identification and characterization of the tolQRA genes of Pseudomonas aeruginosa.J Bacteriol. 1996 Dec;178(24):7059-68. doi: 10.1128/jb.178.24.7059-7068.1996. J Bacteriol. 1996. PMID: 8955385 Free PMC article.
-
TonB/TolA amino-terminal domain modeling.Methods Enzymol. 2007;423:134-48. doi: 10.1016/S0076-6879(07)23005-8. Methods Enzymol. 2007. PMID: 17609129
-
Colicin A binds to a novel binding site of TolA in the Escherichia coli periplasm.Biochem Soc Trans. 2012 Dec 1;40(6):1469-74. doi: 10.1042/BST20120239. Biochem Soc Trans. 2012. PMID: 23176500 Review.
-
TonB-dependent receptors-structural perspectives.Biochim Biophys Acta. 2002 Oct 11;1565(2):318-32. doi: 10.1016/s0005-2736(02)00578-3. Biochim Biophys Acta. 2002. PMID: 12409204 Review.
Cited by
-
Tunable force transduction through the Escherichia coli cell envelope.Proc Natl Acad Sci U S A. 2023 Nov 21;120(47):e2306707120. doi: 10.1073/pnas.2306707120. Epub 2023 Nov 16. Proc Natl Acad Sci U S A. 2023. PMID: 37972066 Free PMC article.
-
Origin and Evolution of Bacterial Periplasmic Force Transducers.Mol Biol Evol. 2025 Jun 4;42(6):msaf138. doi: 10.1093/molbev/msaf138. Mol Biol Evol. 2025. PMID: 40542537 Free PMC article.
-
Structural evidence that colicin A protein binds to a novel binding site of TolA protein in Escherichia coli periplasm.J Biol Chem. 2012 Jun 1;287(23):19048-57. doi: 10.1074/jbc.M112.342246. Epub 2012 Apr 9. J Biol Chem. 2012. PMID: 22493500 Free PMC article.
-
Force-Generation by the Trans-Envelope Tol-Pal System.Front Microbiol. 2022 Mar 3;13:852176. doi: 10.3389/fmicb.2022.852176. eCollection 2022. Front Microbiol. 2022. PMID: 35308353 Free PMC article. Review.
-
In vivo evidence for TonB dimerization.J Bacteriol. 2003 Oct;185(19):5747-54. doi: 10.1128/JB.185.19.5747-5754.2003. J Bacteriol. 2003. PMID: 13129945 Free PMC article.
References
-
- Boulin C., Kempf,R., Koch,M.H.J. and McLaughlin,S.M. (1986) Data appraisal, evaluation and display for synchrotron radiation experiments: hardware and software. Nucl. Instrum. Methods Phys. Res. A, 249, 399–407.
-
- Braun V. and Herrmann,C. (1993) Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol. Microbiol., 8, 261–268. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

