Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs
- PMID: 12169635
- PMCID: PMC126170
- DOI: 10.1093/emboj/cdf432
Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs
Abstract
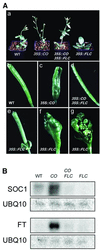
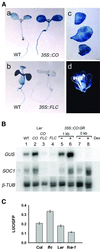
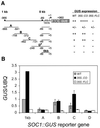
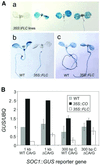
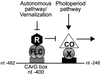
Flowering in Arabidopsis is controlled by endogenous and environmental signals relayed by distinct genetic pathways. The MADS-box flowering-time gene SOC1 is regulated by several pathways and is proposed to co-ordinate responses to environmental signals. SOC1 is directly activated by CONSTANS (CO) in long photoperiods and is repressed by FLC, a component of the vernalization (low-temperature) pathway. We show that in transgenic plants overexpressing CO and FLC, these proteins regulate flowering time antagonistically and FLC blocks transcriptional activation of SOC1 by CO. A series of SOC1::GUS reporter genes identified a 351 bp promoter sequence that mediates activation by CO and repression by FLC. A CArG box (MADS-domain protein binding element) within this sequence was recognized specifically by FLC in vitro and mediated repression by FLC in vivo, suggesting that FLC binds directly to the SOC1 promoter. We propose that CO is recruited to a separate promoter element by a DNA-binding factor and that activation by CO is impaired when FLC is bound to an adjacent CArG motif.
Figures


References
-
- Alvarez-Buylla E.R., Liljegren,S.J., Pelaz,S., Gold,S.E., Burgeff,C., Ditta,G.S., Vergara-Silva,F. and Yanofsky,M.F. (2000) MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J., 24, 457–466. - PubMed
-
- Araki T. (2001) Transition from vegetative to reproductive phase. Curr. Opin. Plant Biol., 4, 63–68. - PubMed
-
- Blázquez M.A. and Weigel,D. (2000) Integration of floral inductive signals in Arabidopsis. Nature, 404, 889–892. - PubMed
-
- Blázquez M.A., Soowal,L.N., Lee,I. and Weigel,D. (1997) LEAFY expression and flower initiation in Arabidopsis. Development, 124, 3835–3844. - PubMed
-
- Borden K.L.B. (1998) RING fingers and B-boxes: zinc-binding protein–protein interaction domains. Biochem. Cell Biol., 76, 351–358. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

