The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis
- PMID: 12169662
- PMCID: PMC123288
- DOI: 10.1073/pnas.162339999
The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis
Abstract
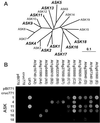
The covalent attachment of ubiquitin is an important determinant for selective protein degradation by the 26S proteasome in plants and animals. The specificity of ubiquitination is often controlled by ubiquitin-protein ligases (or E3s), which facilitate the transfer of ubiquitin to appropriate targets. One ligase type, the SCF E3s are composed of four proteins, cullin1/Cdc53, Rbx1/Roc1/Hrt1, Skp1, and an F-box protein. The F-box protein, which identifies the targets, binds to the Skp1 component of the complex through a degenerate N-terminal approximately 60-aa motif called the F-box. Using published F-boxes as queries, we have identified 694 potential F-box genes in Arabidopsis, making this gene superfamily one of the largest currently known in plants. Most of the encoded proteins contain interaction domains C-terminal to the F-box that presumably participate in substrate recognition. The F-box proteins can be classified via a phylogenetic approach into five major families, which can be further organized into multiple subfamilies. Sequence diversity within the subfamilies suggests that many F-box proteins have distinct functions and/or substrates. Representatives of all of the major families interact in yeast two-hybrid experiments with members of the Arabidopsis Skp family supporting their classification as F-box proteins. For some, a limited preference for Skps was observed, suggesting that a hierarchical organization of SCF complexes exists defined by distinct Skp/F-box protein pairs. Collectively, the data shows that Arabidopsis has exploited the SCF complex and the ubiquitin/26S proteasome pathway as a major route for cellular regulation and that a diverse array of SCF targets is likely present in plants.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

