Effectiveness of capillary electrophoresis using fluorescent-labeled primers in detecting T-cell receptor gamma gene rearrangements
- PMID: 12169674
- PMCID: PMC1906981
- DOI: 10.1016/s1525-1578(10)60694-0
Effectiveness of capillary electrophoresis using fluorescent-labeled primers in detecting T-cell receptor gamma gene rearrangements
Erratum in
- J Mol Diagn. 2003 Aug;5(3):195
Abstract
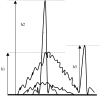
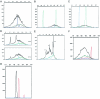
We describe the use of fluorescent-labeled primers to analyze T-cell receptor gamma gene rearrangements (TCR gamma GR) using capillary electrophoresis in the ABI Prism 310 Genetic Analyzer. We also compare the performance with denaturing gradient gel electrophoresis (DGGE). In a single multiplex polymerase chain reaction (PCR) we amplified TCR gamma GR with primers for all known groups of variable region genes, and joining region genes described in lymphoid neoplasms. Ten reactive samples, followed by five cell lines and 25 tumor samples with 41 individual TCR gamma GR (due to many biallelic rearrangements) previously identified by DGGE, were analyzed to validate the technique. The capillary electrophoresis protocol has 92% concordance for both TCR clonal status (23 of 25) and 95% concordance in the number of individual TCR gamma GR (38 of 41) identified by DGGE. The reproducible sensitivity for detecting TCR gamma GR diluted in reactive lymphoid DNA is 2% in clinical applications. Discrimination of predominant rearrangements requires a minimum ratio of two times the height of the normal distribution of polyclonal peaks. Capillary electrophoresis can provide results within 60 minutes for each specimen after PCR is complete. Capillary electrophoresis provides a faster result than sequence-based separation methods and gives an archival electronic record. Fluorescent labeling allows the identification of both the variable and joining gene segments used in a TCR gamma GR. The effectiveness of capillary electrophoresis is similar to DGGE.
Figures
Similar articles
-
Detection of clonal T-cell receptor gamma gene rearrangements in early mycosis fungoides/Sezary syndrome by polymerase chain reaction and denaturing gradient gel electrophoresis (PCR/DGGE).J Invest Dermatol. 1994 Jul;103(1):34-41. doi: 10.1111/1523-1747.ep12389114. J Invest Dermatol. 1994. PMID: 8027579
-
Detection of clonal T-cell receptor gamma gene rearrangements using fluorescent-based PCR and automated high-resolution capillary electrophoresis.Mol Diagn. 2001 Sep;6(3):169-79. doi: 10.1054/modi.2001.27056. Mol Diagn. 2001. PMID: 11571710
-
Analysis of T cell receptor-gamma gene rearrangements by denaturing gradient gel electrophoresis of GC-clamped polymerase chain reaction products. Correlation with tumor-specific sequences.Am J Pathol. 1995 Jan;146(1):46-55. Am J Pathol. 1995. PMID: 7856738 Free PMC article.
-
Polymerase chain reaction/denaturing gradient gel electrophoresis (PCR/DGGE): sensitivity, band pattern analysis, and methodologic optimization.Am J Dermatopathol. 1999 Dec;21(6):547-51. doi: 10.1097/00000372-199912000-00008. Am J Dermatopathol. 1999. PMID: 10608248
-
The distribution of gene segments in T-cell receptor gamma gene rearrangements demonstrates the need for multiple primer sets.J Mol Diagn. 2003 May;5(2):82-7. doi: 10.1016/s1525-1578(10)60456-4. J Mol Diagn. 2003. PMID: 12707372 Free PMC article.
Cited by
-
Results of histopathology, immunohistochemistry, and molecular clonality testing of small intestinal biopsy specimens from clinically healthy client-owned cats.J Vet Intern Med. 2019 Mar;33(2):551-558. doi: 10.1111/jvim.15455. Epub 2019 Feb 28. J Vet Intern Med. 2019. PMID: 30820999 Free PMC article.
-
A New and Simple TRG Multiplex PCR Assay for Assessment of T-cell Clonality: A Comparative Study from the EuroClonality Consortium.Hemasphere. 2019 Jun 4;3(3):e255. doi: 10.1097/HS9.0000000000000255. eCollection 2019 Jun. Hemasphere. 2019. PMID: 31723840 Free PMC article.
-
Cytotoxic peripheral T cell lymphoma arising in a patient with nodular lymphocyte predominant Hodgkin lymphoma: a case report.J Hematop. 2010 Mar 4;3(1):23-8. doi: 10.1007/s12308-010-0055-7. J Hematop. 2010. PMID: 21373174 Free PMC article.
-
Comparison of BIOMED-2 versus laboratory-developed polymerase chain reaction assays for detecting T-cell receptor-gamma gene rearrangements.J Mol Diagn. 2010 Mar;12(2):226-37. doi: 10.2353/jmoldx.2010.090042. J Mol Diagn. 2010. PMID: 20181819 Free PMC article.
-
Validation of a Next-Generation Sequencing-Based T-Cell Receptor Gamma Gene Rearrangement Diagnostic Assay: Transitioning from Capillary Electrophoresis to Next-Generation Sequencing.J Mol Diagn. 2021 Jul;23(7):805-815. doi: 10.1016/j.jmoldx.2021.03.008. Epub 2021 Apr 21. J Mol Diagn. 2021. PMID: 33892183 Free PMC article.
References
-
- Trainor KJ, Brisco MJ, Wan JH, Neoh S, Grist S, Morley AA: Gene rearrangement in B- and T-lymphoproliferative disease detected by the polymerase chain reaction. Blood 1991, 78:192-196 - PubMed
-
- Wood GS, Tung RM, Haeffner AC, Crooks CF, Liao S, Orozco R, Veelken H, Kadin ME, Koh H, Heald P, Uluer AZ: Detection of clonal T-cell receptor γ gene rearrangements in early mycosis fungoides/Sezary syndrome by polymerase chain reaction and denaturing gradient gel electrophoresis (PCR/DGGE). J Invest Dermatol 1994, 103:34-41 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

