Genetic variation for disease resistance and tolerance among Arabidopsis thaliana accessions
- PMID: 12172004
- PMCID: PMC123246
- DOI: 10.1073/pnas.102288999
Genetic variation for disease resistance and tolerance among Arabidopsis thaliana accessions
Abstract
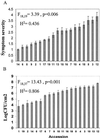
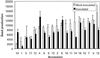
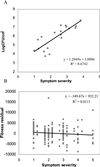
Pathogens can be an important selective agent in plant evolution because they can severely reduce plant fitness and growth. However, the role of pathogen selection on plant evolution depends on the extent of genetic variation for resistance traits and their covariance with host fitness. Although it is usually assumed that resistance traits will covary with plant fitness, this assumption has not been tested rigorously in plant-pathogen interactions. Many plant species are tolerant to herbivores, decoupling the relationship between resistance and fitness. Tolerance to pathogens can reduce selection for resistance and alter the effect of pathogens on plant evolution. In this study, we measured three components of Arabidopsis thaliana resistance (pathogen growth, disease symptoms, and host fitness) to the bacteria Pseudomonas syringae and investigated their covariation to determine the relative importance of resistance and tolerance. We observed extensive quantitative variation in the severity of disease symptoms, the bacterial population size, and the effect of infection on host fitness among 19 accessions of A. thaliana infected with P. syringae. The severity of disease symptoms was strongly and positively correlated with bacterial population size. Although the average fitness of infected plants was smaller than noninfected plants, we found no correlation between the bacterial growth or symptoms expressed by different accessions of A. thaliana and their relative fitness after infection. These results indicate that the accessions studied vary in tolerance to P. syringae, reducing the strength of selection on resistance traits, and that symptoms and bacterial growth are not good predictors of host fitness.
Figures
References
-
- Pimentel D., Lach, L., Zuniga, R. & Morrison, D. (2000) Bioscience 50, 53-65.
-
- Clay K. & Putten, W. H. V. d. (1999) in Life History Evolution in Plants, eds. Vuorisalo, T. & Mutikainen, P. (Kluwer, Dordrecht, The Netherlands), pp. 275–301.
-
- Alexander H. M. (1992) in Plant Resistance to Herbivores and Pathogens, eds. Fritz, R. S. & Simms, E. (Univ. of Chicago Press, Chicago), pp. 326–344.
-
- Leonard K. J. & Levy, Y. (1990) J. Phytopathol. 128, 161-171.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

