High-resolution mapping of the 11q13 amplicon and identification of a gene, TAOS1, that is amplified and overexpressed in oral cancer cells
- PMID: 12172009
- PMCID: PMC123263
- DOI: 10.1073/pnas.172285799
High-resolution mapping of the 11q13 amplicon and identification of a gene, TAOS1, that is amplified and overexpressed in oral cancer cells
Abstract
Amplification of chromosomal band 11q13 is a common event in human cancer. It has been reported in about 45% of head and neck carcinomas and in other cancers including esophageal, breast, liver, lung, and bladder cancer. To understand the mechanism of 11q13 amplification and to identify the potential oncogene(s) driving it, we have fine-mapped the structure of the amplicon in oral squamous cell carcinoma cell lines and localized the proximal and distal breakpoints. A 5-Mb physical map of the region has been prepared from which sequence is available. We quantified copy number of sequence-tagged site markers at 42-550 kb intervals along the length of the amplicon and defined the amplicon core and breakpoints by using TaqMan-based quantitative microsatellite analysis. The core of the amplicon maps to a 1.5-Mb region. The proximal breakpoint localizes to two intervals between sequence-tagged site markers, 550 kb and 160 kb in size, and the distal breakpoint maps to a 250 kb interval. The cyclin D1 gene maps to the amplicon core, as do two new expressed sequence tag clusters. We have analyzed one of these expressed sequence tag clusters and now report that it contains a previously uncharacterized gene, TAOS1 (tumor amplified and overexpressed sequence 1), which is both amplified and overexpressed in oral cancer cells. The data suggest that TAOS1 may be an amplification-dependent candidate oncogene with a role in the development and/or progression of human tumors, including oral squamous cell carcinomas. The approach described here should be useful for characterizing amplified genomic regions in a wide variety of tumors.
Figures

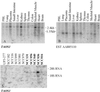

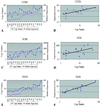
References
-
- Savelyeva L. & Schwab, M. (2001) Cancer Lett. 167, 115-123. - PubMed
-
- Schwab M. (1998) BioEssays 20, 473-479. - PubMed
-
- Åkervall J. A., Jin, Y., Wennerberg, J. P., Zatterstrom, U. K., Kjellen, E., Mertens, F., Willen, R., Mandahl, N., Heim, S. & Mitelman, F. (1995) Cancer 76, 853-859. - PubMed
-
- Gaudray P., Szepetowski, P., Escot, C., Birnbaum, D. & Theillet, C. (1992) Mutat. Res. 276, 317-328. - PubMed
-
- Dickson C., Fantl, V., Gillett, C., Brookes, S., Bartek, J., Smith, R., Fisher, C., Barnes, D. & Peters, G. (1995) Cancer Lett. 90, 43-50. - PubMed
Publication types
MeSH terms
Substances
Associated data
- GDB/D11S5031-5
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

