Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway
- PMID: 12172015
- PMCID: PMC151458
- DOI: 10.1105/tpc.003483
Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway
Abstract
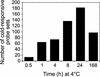
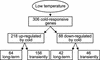
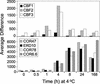
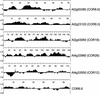
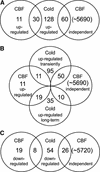
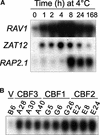
Many plants, including Arabidopsis, increase in freezing tolerance in response to low, nonfreezing temperatures, a phenomenon known as cold acclimation. Previous studies established that cold acclimation involves rapid expression of the CBF transcriptional activators (also known as DREB1 proteins) in response to low temperature followed by induction of the CBF regulon (CBF-targeted genes), which contributes to an increase in freezing tolerance. Here, we present the results of transcriptome-profiling experiments indicating the existence of multiple low-temperature regulatory pathways in addition to the CBF cold response pathway. The transcript levels of approximately 8000 genes were determined at multiple times after plants were transferred from warm to cold temperature and in warm-grown plants that constitutively expressed CBF1, CBF2, or CBF3. A total of 306 genes were identified as being cold responsive, with transcripts for 218 genes increasing and those for 88 genes decreasing threefold or more at one or more time points during the 7-day experiment. These results indicate that extensive downregulation of gene expression occurs during cold acclimation. Of the cold-responsive genes, 48 encode known or putative transcription factors. Two of these, RAP2.1 and RAP2.6, were activated by CBF expression and thus presumably control subregulons of the CBF regulon. Transcriptome comparisons indicated that only 12% of the cold-responsive genes are certain members of the CBF regulon. Moreover, at least 28% of the cold-responsive genes were not regulated by the CBF transcription factors, including 15 encoding known or putative transcription factors, indicating that these cold-responsive genes are members of different low-temperature regulons. Significantly, CBF expression at warm temperatures repressed the expression of eight genes that also were downregulated by low temperature, indicating that in addition to gene induction, gene repression is likely to play an integral role in cold acclimation.
Figures




References
-
- Abler, M.L., and Green, P.L. (1996). Control of mRNA stability in higher plants. Plant Mol. Biol. 32, 63–78. - PubMed
-
- Adamska, I. (1997). ELIPs: Light induced stress proteins. Physiol. Plant. 100, 794–805.
-
- Baker, S.S., Wilhelm, K.S., and Thomashow, M.F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. - PubMed
-
- Ciardi, J.A., Deikman, J., and Orzolek, M.D. (1997). Increased ethylene synthesis enhances chilling tolerance in tomato. Physiol. Plant. 101, 333–340.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

