Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon
- PMID: 12172016
- PMCID: PMC151459
- DOI: 10.1105/tpc.003079
Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon
Abstract
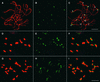
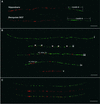
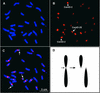
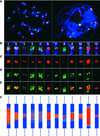
The centromere of eukaryotic chromosomes is essential for the faithful segregation and inheritance of genetic information. In the majority of eukaryotic species, centromeres are associated with highly repetitive DNA, and as a consequence, the boundary for a functional centromere is difficult to define. In this study, we demonstrate that the centers of rice centromeres are occupied by a 155-bp satellite repeat, CentO, and a centromere-specific retrotransposon, CRR. The CentO satellite is located within the chromosomal regions to which the spindle fibers attach. CentO is quantitatively variable among the 12 rice centromeres, ranging from 65 kb to 2 Mb, and is interrupted irregularly by CRR elements. The break points of 14 rice centromere misdivision events were mapped to the middle of the CentO arrays, suggesting that the CentO satellite is located within the functional domain of rice centromeres. Our results demonstrate that the CentO satellite may be a key DNA element for rice centromere function.
Figures


References
-
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Aragon-Alcaide, L., Miller, T., Schwarzacher, T., Reader, S., and Moore, G. (1996). A cereal centromeric sequence. Chromosoma 105, 261–268. - PubMed
-
- Arumuganathan, K., and Earle, E.D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–218.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

