FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis
- PMID: 12172022
- PMCID: PMC151465
- DOI: 10.1105/tpc.001495
FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis
Abstract
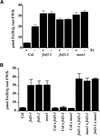
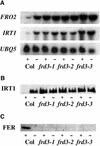
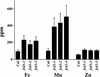
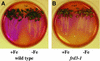
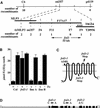
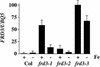
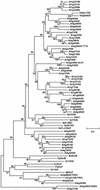
We present the cloning and characterization of an Arabidopsis gene, FRD3, involved in iron homeostasis. Plants carrying any of the three alleles of frd3 constitutively express three strategy I iron deficiency responses and misexpress a number of iron deficiency-regulated genes. Mutant plants also accumulate approximately twofold excess iron, fourfold excess manganese, and twofold excess zinc in their shoots. frd3-3 was first identified as man1. The FRD3 gene is expressed at detectable levels in roots but not in shoots and is predicted to encode a membrane protein belonging to the multidrug and toxin efflux family. Other members of this family have been implicated in a variety of processes and are likely to transport small organic molecules. The phenotypes of frd3 mutant plants, which are consistent with a defect in either iron deficiency signaling or iron distribution, indicate that FRD3 is an important component of iron homeostasis in Arabidopsis.
Figures

Similar articles
-
The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation.Plant Physiol. 2007 May;144(1):197-205. doi: 10.1104/pp.107.097162. Epub 2007 Mar 9. Plant Physiol. 2007. PMID: 17351051 Free PMC article.
-
Natural variation at the FRD3 MATE transporter locus reveals cross-talk between Fe homeostasis and Zn tolerance in Arabidopsis thaliana.PLoS Genet. 2012;8(12):e1003120. doi: 10.1371/journal.pgen.1003120. Epub 2012 Dec 6. PLoS Genet. 2012. PMID: 23236296 Free PMC article.
-
Intertwined metal homeostasis, oxidative and biotic stress responses in the Arabidopsis frd3 mutant.Plant J. 2020 Apr;102(1):34-52. doi: 10.1111/tpj.14610. Epub 2020 Jan 4. Plant J. 2020. PMID: 31721347
-
The zinc homeostasis network of land plants.Biochim Biophys Acta. 2012 Sep;1823(9):1553-67. doi: 10.1016/j.bbamcr.2012.05.016. Epub 2012 May 22. Biochim Biophys Acta. 2012. PMID: 22626733 Review.
-
Getting a sense for signals: regulation of the plant iron deficiency response.Biochim Biophys Acta. 2012 Sep;1823(9):1521-30. doi: 10.1016/j.bbamcr.2012.03.010. Epub 2012 Mar 28. Biochim Biophys Acta. 2012. PMID: 22483849 Free PMC article. Review.
Cited by
-
Characterization of Al-responsive citrate excretion and citrate-transporting MATEs in Eucalyptus camaldulensis.Planta. 2013 Apr;237(4):979-89. doi: 10.1007/s00425-012-1810-z. Epub 2012 Nov 28. Planta. 2013. PMID: 23187679
-
Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes.BMC Plant Biol. 2006 Oct 12;6:25. doi: 10.1186/1471-2229-6-25. BMC Plant Biol. 2006. PMID: 17038189 Free PMC article.
-
Microarray analysis of iron deficiency chlorosis in near-isogenic soybean lines.BMC Genomics. 2007 Dec 21;8:476. doi: 10.1186/1471-2164-8-476. BMC Genomics. 2007. PMID: 18154662 Free PMC article.
-
Identification of two chickpea multidrug and toxic compound extrusion transporter genes transcriptionally upregulated upon aluminum treatment in root tips.Front Plant Sci. 2022 Aug 5;13:909045. doi: 10.3389/fpls.2022.909045. eCollection 2022. Front Plant Sci. 2022. PMID: 35991422 Free PMC article.
-
MYB10 and MYB72 are required for growth under iron-limiting conditions.PLoS Genet. 2013 Nov;9(11):e1003953. doi: 10.1371/journal.pgen.1003953. Epub 2013 Nov 21. PLoS Genet. 2013. PMID: 24278034 Free PMC article.
References
-
- Alonso, J.M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284, 2148–2152. - PubMed
-
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Askwith, C., and Kaplan, J. (1998). Iron and copper transport in yeast and its relevance to human disease. Trends Biochem. Sci. 23, 135–138. - PubMed
-
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (2002). Current Protocols in Molecular Biology. (New York: John Wiley & Sons).
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

