Large-scale proteomic analysis of the human spliceosome
- PMID: 12176931
- PMCID: PMC186633
- DOI: 10.1101/gr.473902
Large-scale proteomic analysis of the human spliceosome
Abstract
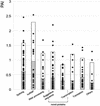
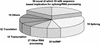
In a previous proteomic study of the human spliceosome, we identified 42 spliceosome-associated factors, including 19 novel ones. Using enhanced mass spectrometric tools and improved databases, we now report identification of 311 proteins that copurify with splicing complexes assembled on two separate pre-mRNAs. All known essential human splicing factors were found, and 96 novel proteins were identified, of which 55 contain domains directly linking them to functions in splicing/RNA processing. We also detected 20 proteins related to transcription, which indicates a direct connection between this process and splicing. This investigation provides the most detailed inventory of human spliceosome-associated factors to date, and the data indicate a number of interesting links coordinating splicing with other steps in the gene expression pathway.
Figures


References
-
- Ajuh P, Sleeman J, Chusainow J, Lamond AI. A direct interaction between the carboxyl-terminal region of CDC5L and the WD40 domain of PLRG1 is essential for pre-mRNA splicing. J Biol Chem. 2001;276:42370–42381. - PubMed
-
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002a;12:1–11. - PubMed
-
- Andersen, J.S., Rappsilber, J., Ishihama, Y., Wilkins, C., Nigg, E., and Mann, M. 2002b. “LC MS/MS of complex peptide mixtures using the pulsing feature of the hybrid QSTAR-pulsar quadrupole time-of-flight mass spectrometer to increase protein identification.” Paper presented at The 50th ASMS Conference on Mass Spectrometry and Allied Topics (Orlando, FL, USA).
-
- Aravind L, Koonin EV. G-patch: A new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem Sci. 1999;24:342–344. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
