Voluntary ethanol consumption by mice: genome-wide analysis of quantitative trait loci and their interactions in a C57BL/6ByJ x 129P3/J F2 intercross
- PMID: 12176933
- PMCID: PMC186641
- DOI: 10.1101/gr.129702
Voluntary ethanol consumption by mice: genome-wide analysis of quantitative trait loci and their interactions in a C57BL/6ByJ x 129P3/J F2 intercross
Abstract
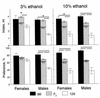
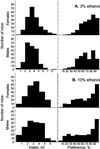
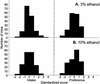
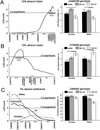
Consumption of ethanol solutions by rodents in two-bottle choice tests is a model to study human alcohol intake. Mice of the C57BL/6ByJ strain have higher ethanol preferences and intakes than do mice of the 129P3/J strain. F2 hybrids between these two strains were phenotyped using two-bottle tests involving a choice between water and either 3% or 10% ethanol. High ethanol preferences and intakes of the B6 mice were inherited as additive or dominant traits in the F2 generation. A genome screen using these F2 mice identified three significant linkages. Two loci, on distal chromosome 4 (Ap3q) and proximal chromosome 7 (Ap7q), strongly affected 10% ethanol intake and weakly affected 3% ethanol intake. A male-specific locus on proximal chromosome 8 (Ap8q) affected 3% ethanol preference, but not indexes of 10% ethanol consumption. In addition, six suggestive linkages (on chromosomes 2, 9, 12, 13, 17, and 18) affecting indexes of 3% and/or 10% ethanol consumption were detected. The loci with significant and suggestive linkages accounted for 35-44% of the genetic variation in ethanol consumption phenotypes. No additive-by-additive epistatic interactions were detected for the primary loci with significant and suggestive linkages. However, there were a few modifiers of the primary linkages and a number of interactions among unlinked loci. This demonstrates a significant role of the genetic background in the variation of ethanol consumption.
Figures

References
-
- Arola L, Roig R, Cascon E, Brunet MJ, Fornos N, Sabate M, Raga X, Batista J, Salvado MJ, Blade C. Model for voluntary wine and alcohol consumption in rats. Physiol Behav. 1997;62:353–357. - PubMed
-
- Bachmanov AA, Beauchamp GK, Tordoff MG. Voluntary consumption of ethanol in 28 inbred mouse strains (Abstract) Alcohol Clin Exp Res. 2000;24:58A. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
