FGF signaling targets the pRb-related p107 and p130 proteins to induce chondrocyte growth arrest
- PMID: 12177046
- PMCID: PMC2174007
- DOI: 10.1083/jcb.200205025
FGF signaling targets the pRb-related p107 and p130 proteins to induce chondrocyte growth arrest
Abstract
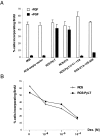
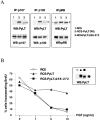
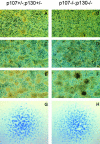
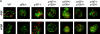
Unregulated FGF signaling affects endochondral ossification and long bone growth, causing several genetic forms of human dwarfism. One major mechanism by which FGFs regulate endochondral bone growth is through their inhibitory effect on chondrocyte proliferation. Because mice with targeted mutations of the retinoblastoma (Rb)-related proteins p107 and p130 present severe endochondral bone defects with excessive chondrocyte proliferation, we have investigated the role of the Rb family of cell cycle regulators in the FGF response. Using a chondrocyte cell line, we found that FGF induced a rapid dephosphorylation of all three proteins of the Rb family (pRb, p107, and p130) and a blockade of the cells in the G1 phase of the cell cycle. This cell cycle block was reversed by inactivation of Rb proteins with viral oncoproteins such as polyoma large T (PyLT) antigen and Adenovirus E1A. Expression of a PyLT mutant that efficiently binds pRb, but not p107 and p130, allowed the cells to be growth inhibited by FGF, suggesting that pRb itself is not involved in the FGF response. To investigate more precisely the role of the individual Rb family proteins in FGF-mediated growth inhibition, we used chondrocyte micromass culture of limb bud cells isolated from mice lacking Rb proteins individually or in combination. Although wild-type as well as Rb-/- chondrocytes were similarly growth inhibited by FGF, chondrocytes null for p107 and p130 did not respond to FGF. Furthermore, FGF treatment of metatarsal bone rudiments obtained from p107-/-;p130-/- embryos failed to inhibit proliferation of growth plate chondrocytes, whereas rudiments from p107-null or p130-null embryos showed only a slight inhibition of growth. Our findings indicate that p107 and p130, but not pRb, are critical effectors of FGF-mediated growth inhibition in chondrocytes.
Figures





Similar articles
-
Shared role of the pRB-related p130 and p107 proteins in limb development.Genes Dev. 1996 Jul 1;10(13):1633-44. doi: 10.1101/gad.10.13.1633. Genes Dev. 1996. PMID: 8682294
-
Expression of the retinoblastoma family of tumor suppressors during murine embryonic orofacial development.Orthod Craniofac Res. 2003 Feb;6(1):32-47. doi: 10.1046/j.1439-0280.2003.2c035.x. Orthod Craniofac Res. 2003. PMID: 12627794
-
p107 and p130 Coordinately regulate proliferation, Cbfa1 expression, and hypertrophic differentiation during endochondral bone development.Dev Biol. 2002 Jul 15;247(2):271-85. doi: 10.1006/dbio.2002.0691. Dev Biol. 2002. PMID: 12086466
-
p107 and p130: versatile proteins with interesting pockets.Exp Cell Res. 2001 Mar 10;264(1):135-47. doi: 10.1006/excr.2000.5135. Exp Cell Res. 2001. PMID: 11237530 Review.
-
Activity of the retinoblastoma family proteins, pRB, p107, and p130, during cellular proliferation and differentiation.Crit Rev Biochem Mol Biol. 1996 Jun;31(3):237-71. doi: 10.3109/10409239609106585. Crit Rev Biochem Mol Biol. 1996. PMID: 8817077 Review.
Cited by
-
PP2A: more than a reset switch to activate pRB proteins during the cell cycle and in response to signaling cues.Cell Cycle. 2015;14(1):18-30. doi: 10.4161/15384101.2014.985069. Cell Cycle. 2015. PMID: 25483052 Free PMC article. Review.
-
Differential gene expression of the intermediate and outer interzone layers of developing articular cartilage in murine embryos.Stem Cells Dev. 2014 Aug 15;23(16):1883-98. doi: 10.1089/scd.2013.0235. Epub 2014 Jun 16. Stem Cells Dev. 2014. PMID: 24738827 Free PMC article.
-
FGFR3 signaling induces a reversible senescence phenotype in chondrocytes similar to oncogene-induced premature senescence.Bone. 2010 Jul;47(1):102-10. doi: 10.1016/j.bone.2010.03.021. Epub 2010 Mar 31. Bone. 2010. PMID: 20362703 Free PMC article.
-
MPSI Manifestations and Treatment Outcome: Skeletal Focus.Int J Mol Sci. 2022 Sep 22;23(19):11168. doi: 10.3390/ijms231911168. Int J Mol Sci. 2022. PMID: 36232472 Free PMC article. Review.
-
Cell type-specific control of protein synthesis and proliferation by FGF-dependent signaling to the translation repressor 4E-BP.Proc Natl Acad Sci U S A. 2016 Jul 5;113(27):7545-50. doi: 10.1073/pnas.1605451113. Epub 2016 Jun 16. Proc Natl Acad Sci U S A. 2016. PMID: 27313212 Free PMC article.
References
-
- Ahrens, P.B., M. Solursh, R.S. Reiter, and C.T. Singley. 1979. Position-related capacity for differentiation of limb mesenchyme in cell culture. Dev. Biol. 69:436–450. - PubMed
-
- Aikawa, T., G.V. Segre, and K. Lee. 2001. Fibroblast growth factor inhibits chondrocytic growth through induction of p21 and subsequent inactivation of cyclin E-Cdk2. J. Biol. Chem. 276:29347–29352. - PubMed
-
- Basilico, C., and D. Moscatelli. 1992. The FGF family of growth factors and oncogenes. Adv. Cancer Res. 59:115–165. - PubMed
-
- Clarke, A.R., E.R. Maandag, M. van Roon, N.M. van der Lugt, M. van der Valk, M.L. Hooper, A. Berns, and H. te Riele. 1992. Requirement for a functional Rb-1 gene in murine development. Nature. 359:328–330. - PubMed
-
- Classon, M., and N. Dyson. 2001. p107 and p130: versatile proteins with interesting pockets. Exp. Cell Res. 264:135–147. - PubMed

