Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family
- PMID: 12177301
- PMCID: PMC134238
- DOI: 10.1093/nar/gkf470
Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family
Abstract
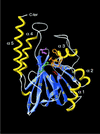
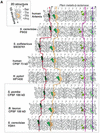
A separate family of enzymes within the metallo-beta-lactamase fold comprises several important proteins acting on nucleic acid substrates, involved in DNA repair (Artemis, SNM1 and PSO2) and RNA processing [cleavage and polyadenylation specificity factor (CPSF) subunit]. Proteins of this family, named beta-CASP after the names of its representative members, possess specific features relative to those of other metallo-beta-lactamases, that are concentrated in the C-terminal part of the domain. In this study, using sensitive methods of sequence analysis, we identified highly conserved amino acids specific to the beta-CASP family, some of which were unidentified to date, that are predicted to play critical roles in the enzymatic function. The identification and characterisation of all the extant, detectable beta-CASP members within sequence databases and genome data also allowed us to unravel particular sequence features which are likely to be involved in substrate specificity, as well as to describe new but as yet uncharacterised members which may play critical roles in DNA and RNA metabolism.
Figures

References
-
- Aravind L. (1999) An evolutionary classification of the metallo-β-lactamase fold proteins. In Silico Biol., 1, 69–91. - PubMed
-
- Melino S., Capo,C., Dragani,B., Aceto,A. and Petruzzelli,R. (1998) A zinc-binding motif conserved in glyoxalase II, β-lactamase and arylsulfatases. Trends Biochem. Sci., 23, 381–382. - PubMed
-
- Daiyasu H., Osaka,K., Ishino,Y. and Toh,H. (2001) Expansion of the zinc metallo-hydrolase family of the β-lactamase fold. FEBS Lett., 503, 1–6. - PubMed
-
- Moshous D., Callebaut,I., de Chasseval,R., Corneo,B., Cavazzana-Calvo,M., Le Deist,F., Tezcan,I., Sanal,O., Bertrand,Y., Philippe,N. et al. (2001) ARTEMIS, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell, 105, 177–186. - PubMed
-
- Ma Y., Pannicke,U., Schwarz,K. and Lieber,M.R. (2002) Hairpin opening and overhang processing by an Artemis/DNA-dependent complex in non-homologous end joining and V(D)J recombination. Cell, 108, 781–794. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

