The human XPC DNA repair gene: arrangement, splice site information content and influence of a single nucleotide polymorphism in a splice acceptor site on alternative splicing and function
- PMID: 12177305
- PMCID: PMC134237
- DOI: 10.1093/nar/gkf469
The human XPC DNA repair gene: arrangement, splice site information content and influence of a single nucleotide polymorphism in a splice acceptor site on alternative splicing and function
Abstract
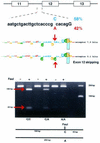
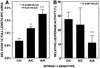
XPC DNA repair gene mutations result in the cancer-prone disorder xeroderma pigmentosum. The XPC gene spans 33 kb and has 16 exons (82-882 bp) and 15 introns (0.08-5.4 kb). A 1.6 kb intron was found within exon 5. Sensitive real- time quantitative reverse transcription-polymerase chain reaction methods were developed to measure full-length XPC mRNA (the predominant form) and isoforms that skipped exons 4, 7 or 12. Exon 7 was skipped in approximately 0.07% of XPC mRNAs, consistent with the high information content of the exon 7 splice acceptor and donor sites (12.3 and 10.4 bits). In contrast, exon 4 was skipped in approximately 0.7% of the XPC mRNAs, consistent with the low information content of the exon 4 splice acceptor (-0.1 bits). A new common C/A single nucleotide polymorphism in the XPC intron 11 splice acceptor site (58% C in 97 normals) decreased its information content from 7.5 to 5.1 bits. Fibroblasts homozygous for A/A had significantly higher levels (approximately 2.6-fold) of the XPC mRNA isoform that skipped exon 12 than those homozygous for C/C. This abnormally spliced XPC mRNA isoform has diminished DNA repair function and may contribute to cancer susceptibility.
Figures



References
-
- Van Steeg H. and Kraemer,K.H. (1999) Xeroderma pigmentosum and the role of UV-induced DNA damage in skin cancer. Mol. Med. Today, 5, 86–94. - PubMed
-
- Kraemer K.H. (1999) Heritable diseases with increased sensitivity to cellular injury. In Freedberg,I.M., Eisen,A.Z., Wolff,K., Austen,K.F., Goldsmith,L., Katz,S.I. and Fitzpatrick,T.B. (eds), Fitzpatrick’s Dermatology in General Medicine. McGraw Hill, New York, NY, pp. 1848–1862.
-
- Kraemer K.H., Lee,M.M. and Scotto,J. (1987) Xeroderma pigmentosum. Cutaneous, ocular and neurologic abnormalities in 830 published cases. Arch. Dermatol., 123, 241–250. - PubMed
-
- Kraemer K.H., Lee,M.-M., Andrews,A.D. and Lambert,W.C. (1994) The role of sunlight and DNA repair in melanoma and nonmelanoma skin cancer: the xeroderma pigmentosum paradigm. Arch. Dermatol., 130, 1018–1021. - PubMed
-
- Legerski R. and Peterson,C. (1992) Expression cloning of a human DNA repair gene involved in xeroderma pigmentosum group C. Nature, 359, 70–73. [Erratum (1992) Nature, 360, 610.] - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

