Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags
- PMID: 12177431
- PMCID: PMC129300
- DOI: 10.1073/pnas.172170199
Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags
Abstract
Understanding biological systems and the roles of their constituents is facilitated by the ability to make quantitative, sensitive, and comprehensive measurements of how their proteome changes, e.g., in response to environmental perturbations. To this end, we have developed a high-throughput methodology to characterize an organism's dynamic proteome based on the combination of global enzymatic digestion, high-resolution liquid chromatographic separations, and analysis by Fourier transform ion cyclotron resonance mass spectrometry. The peptides produced serve as accurate mass tags for the proteins and have been used to identify with high confidence >61% of the predicted proteome for the ionizing radiation-resistant bacterium Deinococcus radiodurans. This fraction represents the broadest proteome coverage for any organism to date and includes 715 proteins previously annotated as either hypothetical or conserved hypothetical.
Figures
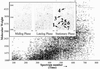

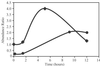
Comment in
-
New technology may reveal mechanisms of radiation resistance in Deinococcus radiodurans.Proc Natl Acad Sci U S A. 2002 Aug 20;99(17):10943-4. doi: 10.1073/pnas.182429699. Epub 2002 Aug 12. Proc Natl Acad Sci U S A. 2002. PMID: 12177454 Free PMC article. No abstract available.
References
-
- Venter J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304-1325. - PubMed
-
- Daly M. J. (2000) Biotechnology 11, 280-285. - PubMed
-
- Smith K. C. & Martignoni, K. D. (1976) Photochem. Photobiol. 24, 515-523. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

