Expression and molecular analysis of the Arabidopsis DXR gene encoding 1-deoxy-D-xylulose 5-phosphate reductoisomerase, the first committed enzyme of the 2-C-methyl-D-erythritol 4-phosphate pathway
- PMID: 12177470
- PMCID: PMC166745
- DOI: 10.1104/pp.003798
Expression and molecular analysis of the Arabidopsis DXR gene encoding 1-deoxy-D-xylulose 5-phosphate reductoisomerase, the first committed enzyme of the 2-C-methyl-D-erythritol 4-phosphate pathway
Abstract
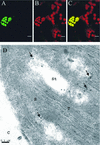
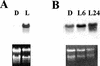
1-Deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) catalyzes the first committed step of the 2-C-methyl-D-erythritol 4-phosphate pathway for isoprenoid biosynthesis. In Arabidopsis, DXR is encoded by a single-copy gene. We have cloned a full-length cDNA corresponding to this gene. A comparative analysis of all plant DXR sequences known to date predicted an N-terminal transit peptide for plastids, with a conserved cleavage site, and a conserved proline-rich region at the N terminus of the mature protein, which is not present in the prokaryotic DXR homologs. We demonstrate that Arabidopsis DXR is targeted to plastids and localizes into chloroplasts of leaf cells. The presence of the proline-rich region in the mature Arabidopsis DXR was confirmed by detection with a specific antibody. A proof of the enzymatic function of this protein was obtained by complementation of an Escherichia coli mutant defective in DXR activity. The expression pattern of beta-glucuronidase, driven by the DXR promoter in Arabidopsis transgenic plants, together with the tissue distribution of DXR transcript and protein, revealed developmental and environmental regulation of the DXR gene. The expression pattern of the DXR gene parallels that of the Arabidopsis 1-deoxy-D-xylulose 5-phosphate synthase gene, but the former is slightly more restricted. These genes are expressed in most organs of the plant including roots, with higher levels in seedlings and inflorescences. The block of the 2-C-methyl-D-erythritol 4-phosphate pathway in Arabidopsis seedlings with fosmidomycin led to a rapid accumulation of DXR protein, whereas the 1-deoxy-D-xylulose 5-phosphate synthase protein level was not altered. Our results are consistent with the participation of the Arabidopsis DXR gene in the control of the 2-C-methyl-D-erythritol 4-phosphate pathway.
Figures





References
-
- Araus JL, Bort J, Brown RH, Bassett CL, Cortadellas N. Immunocytochemical localization of phosphoenolpyruvte carboxylase and photosynthetic gas-exchange characteristics in ears of Triticum durum Desf. Planta. 1993;191:507–514.
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley and Sons, Inc.; 1987.
-
- Axelos M, Curei C, Mazzolini L, Bardet C, Lescure B. A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol Biochem. 1992;30:123–128.
-
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

