The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis
- PMID: 12177480
- PMCID: PMC166755
- DOI: 10.1104/pp.003418
The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis
Abstract
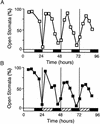
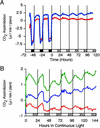
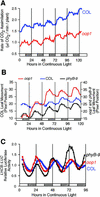
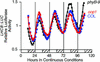
Arabidopsis displays circadian rhythms in stomatal aperture, stomatal conductance, and CO(2) assimilation, each of which peaks around the middle of the day. The rhythmic opening and closing of stomata confers a rhythm in sensitivity and resistance, respectively, to the toxic gas sulfur dioxide. Using this physiological assay as a basis for a mutant screen, we isolated mutants with defects in circadian timing. Here, we characterize one mutant, out of phase 1 (oop1), with the circadian phenotype of altered phase. That is, the timing of the peak (acrophase) of multiple circadian rhythms (leaf movement, CO(2) assimilation, and LIGHT-HARVESTING CHLOROPHYLL a/b-BINDING PROTEIN transcription) is early with respect to wild type, although all circadian rhythms retain normal period length. This is the first such mutant to be characterized in Arabidopsis. oop1 also displays a strong photoperception defect in red light characteristic of phytochrome B (phyB) mutants. The oop1 mutation is a nonsense mutation of PHYB that results in a truncated protein of 904 amino acids. The defect in circadian phasing is seen in seedlings entrained by a light-dark cycle but not in seedlings entrained by a temperature cycle. Thus, PHYB contributes light information critical for proper determination of circadian phase.
Figures



References
-
- Ahmad M, Jarillo JA, Smirnova O, Cashmore AR. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell. 1998;1:939–948. - PubMed
-
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. - PubMed
-
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
-
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

