A new resource of locally transposed Dissociation elements for screening gene-knockout lines in silico on the Arabidopsis genome
- PMID: 12177482
- PMCID: PMC166757
- DOI: 10.1104/pp.002774
A new resource of locally transposed Dissociation elements for screening gene-knockout lines in silico on the Arabidopsis genome
Abstract
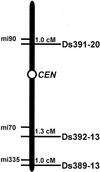
We transposed Dissociation (Ds) elements from three start loci on chromosome 5 in Arabidopsis (Nossen ecotype) by using a local transposition system. We determined partial genomic sequences flanking the Ds elements and mapped the elements' insertion sites in 1,173 transposed lines by comparison with the published genomic sequence. Most of the lines contained a single copy of the Ds element. One-half of the lines contained Ds on chromosome 5; in particular, insertion "hot spots" near the three start loci were clearly observed. In the other lines, the Ds elements were transposed across chromosomes. We found other insertion hot spots at the tops of chromosomes 2 and 4, near nucleolus organizer regions 2 and 4, respectively. Another characteristic feature was that the Ds elements tended to transpose near the chromosome ends and rarely transposed near centromeres. The distribution patterns differed among the three start loci, even though they possessed the same Ds construct. More than one-half of the Ds elements were inserted irregularly into the genome; that is, they did not retain the perfect inverted repeat sequence of Ds nor leave perfect target site duplications. This precise analysis of distribution patterns will contribute to a comprehensive understanding of the transposing mechanism. From these Ds insertion sites, we have constructed a database for screening gene-knockout mutants in silico. In 583 of the 1,173 lines, the Ds elements were inserted into protein-coding genes, which suggests that these lines are gene-knockout mutants. The database and individual lines will be available freely for academic use from the RIKEN Bio-Resource Center (http://www.brc.riken.go.jp/Eng/index.html).
Figures


References
-
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
-
- Azpiroz-Leehan R, Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet. 1997;13:152–156. - PubMed
-
- Fedoroff NV, Smith DL. A versatile system for detecting transposition in Arabidopsis. Plant J. 1993;3:273–289. - PubMed
-
- Ito T, Kim G-T, Shinozaki K. Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J. 2000;22:257–264. - PubMed
-
- Ito T, Motohashi R, Shinozaki K. Preparation of transposon insertion lines and determination of the insertion sites in Arabidopsis genome. In: Braman J, editor. Methods in Molecular Biology. Ed 2. 182, In Vitro Mutagenesis Protocols. Totowa, NJ: Humana Press; 2001. pp. 211–221. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

