Fatty acid export from the chloroplast. Molecular characterization of a major plastidial acyl-coenzyme A synthetase from Arabidopsis
- PMID: 12177483
- PMCID: PMC166758
- DOI: 10.1104/pp.003251
Fatty acid export from the chloroplast. Molecular characterization of a major plastidial acyl-coenzyme A synthetase from Arabidopsis
Abstract
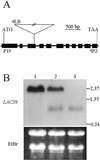
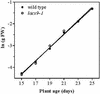
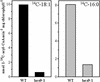
Acyl-coenzyme A (CoA) synthetases (ACSs, EC 6.2.1.3) catalyze the formation of fatty acyl-CoAs from free fatty acid, ATP, and CoA. Essentially all de novo fatty acid synthesis occurs in the plastid. Fatty acids destined for membrane glycerolipid and triacylglycerol synthesis in the endoplasmic reticulum must be first activated to acyl-CoAs via an ACS. Within a family of nine ACS genes from Arabidopsis, we identified a chloroplast isoform, LACS9. LACS9 is highly expressed in developing seeds and young rosette leaves. Both in vitro chloroplast import assays and transient expression of a green fluorescent protein fusion indicated that the LACS9 protein is localized in the plastid envelope. A T-DNA knockout mutant (lacs9-1) was identified by reverse genetics and these mutant plants were indistinguishable from wild type in growth and appearance. Analysis of leaf lipids provided no evidence for compromised export of acyl groups from chloroplasts. However, direct assays demonstrated that lacs9-1 plants contained only 10% of the chloroplast long-chain ACS activity found for wild type. The residual long-chain ACS activity in mutant chloroplasts was comparable with calculated rates of fatty acid synthesis. Although another isozyme contributes to the activation of fatty acids during their export from the chloroplast, LACS9 is a major chloroplast ACS.
Figures


Similar articles
-
Insertional mutant analysis reveals that long-chain acyl-CoA synthetase 1 (LACS1), but not LACS8, functionally overlaps with LACS9 in Arabidopsis seed oil biosynthesis.Plant J. 2010 Dec;64(6):1048-58. doi: 10.1111/j.1365-313X.2010.04396.x. Epub 2010 Nov 15. Plant J. 2010. PMID: 21143684
-
Two activities of long-chain acyl-coenzyme A synthetase are involved in lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis.Plant Physiol. 2015 Feb;167(2):351-66. doi: 10.1104/pp.114.250365. Epub 2014 Dec 24. Plant Physiol. 2015. PMID: 25540329 Free PMC article.
-
Functional Analysis of Rice Long-Chain Acyl-CoA Synthetase 9 (OsLACS9) in the Chloroplast Envelope Membrane.Int J Mol Sci. 2020 Mar 23;21(6):2223. doi: 10.3390/ijms21062223. Int J Mol Sci. 2020. PMID: 32210132 Free PMC article.
-
Yeast acyl-CoA synthetases at the crossroads of fatty acid metabolism and regulation.Biochim Biophys Acta. 2007 Mar;1771(3):286-98. doi: 10.1016/j.bbalip.2006.05.003. Epub 2006 May 16. Biochim Biophys Acta. 2007. PMID: 16798075 Review.
-
An Arabidopsis family of six acyl-CoA-binding proteins has three cytosolic members.Plant Physiol Biochem. 2009 Jun;47(6):479-84. doi: 10.1016/j.plaphy.2008.12.002. Epub 2008 Dec 16. Plant Physiol Biochem. 2009. PMID: 19121948 Review.
Cited by
-
Identification of hydroxy fatty acid and triacylglycerol metabolism-related genes in lesquerella through seed transcriptome analysis.BMC Genomics. 2015 Mar 24;16(1):230. doi: 10.1186/s12864-015-1413-8. BMC Genomics. 2015. PMID: 25881190 Free PMC article.
-
The plant cell wall matrix harbors a precursor of defense signaling peptides.Proc Natl Acad Sci U S A. 2005 Sep 6;102(36):12974-7. doi: 10.1073/pnas.0505248102. Epub 2005 Aug 26. Proc Natl Acad Sci U S A. 2005. PMID: 16126900 Free PMC article.
-
Molecular characterization of an Arabidopsis acyl-coenzyme a synthetase localized on glyoxysomal membranes.Plant Physiol. 2002 Dec;130(4):2019-26. doi: 10.1104/pp.012955. Plant Physiol. 2002. PMID: 12481085 Free PMC article.
-
A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis.Plant Cell. 2009 Feb;21(2):507-25. doi: 10.1105/tpc.108.062513. Epub 2009 Feb 13. Plant Cell. 2009. PMID: 19218397 Free PMC article.
-
Fluorescent Protein Aided Insights on Plastids and their Extensions: A Critical Appraisal.Front Plant Sci. 2016 Jan 20;6:1253. doi: 10.3389/fpls.2015.01253. eCollection 2015. Front Plant Sci. 2016. PMID: 26834765 Free PMC article. Review.
References
-
- Bao X, Focke M, Pollard M, Ohlrogge J. Understanding in vivo carbon precursor supply for fatty acid synthesis in leaf tissue. Plant J. 2000;22:39–50. - PubMed
-
- Black PN, DiRusso CC, Metzger AK, Heimert TL. Cloning, sequencing, and expression of the fadD gene of Escherichia coli encoding acyl coenzyme A synthetase. J Biol Chem. 1992;267:25513–25520. - PubMed
-
- Black PN, DiRusso CC, Sherin D, MacColl R, Knudsen J, Weimar JD. Affinity labeling fatty acyl-CoA synthetase with 9-p-azidophenoxy nonanoic acid and the identification of the fatty acid-binding site. J Biol Chem. 2000;275:38547–38553. - PubMed
-
- Black PN, Zhang Q, Weimar JD, DiRusso CC. Mutational analysis of a fatty acyl-coenzyme A synthetase signature motif identifies seven amino acid residues that modulate fatty acid substrate specificity. J Biol Chem. 1997;272:4896–4903. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

