Crp79p, like Mex67p, is an auxiliary mRNA export factor in Schizosaccharomyces pombe
- PMID: 12181330
- PMCID: PMC117926
- DOI: 10.1091/mbc.e01-11-0133
Crp79p, like Mex67p, is an auxiliary mRNA export factor in Schizosaccharomyces pombe
Abstract
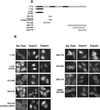
The export of mRNA from the nucleus to the cytoplasm involves interactions of proteins with mRNA and the nuclear pore complex. We isolated Crp79p, a novel mRNA export factor from the same synthetic lethal screen that led to the identification of spMex67p in Schizosaccharomyces pombe. Crp79p is a 710-amino-acid-long protein that contains three RNA recognition motif domains in tandem and a distinct C-terminus. Fused to green fluorescent protein (GFP), Crp79p localizes to the cytoplasm. Like Mex67p, Crp79-GFP binds poly(A)(+) RNA in vivo, shuttles between the nucleus and the cytoplasm, and contains a nuclear export activity at the C-terminus that is Crm1p-independent. All of these properties are essential for Crp79p to promote mRNA export. Crp79p import into the nucleus depends on the Ran system. A domain of spMex67p previously identified as having a nuclear export activity can functionally substitute for the nuclear export activity at the C-terminus of Crp79p. Although both Crp79p and spMex67p function to export mRNA, Crp79p does not substitute for all of spMex67p functions and probably is not a functional homologue of spMex67p. We propose that Crp79p is a nonessential mRNA export carrier in S. pombe.
Figures


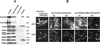



Similar articles
-
Mex67p of Schizosaccharomyces pombe interacts with Rae1p in mediating mRNA export.Mol Cell Biol. 2000 Dec;20(23):8767-82. doi: 10.1128/MCB.20.23.8767-8782.2000. Mol Cell Biol. 2000. PMID: 11073978 Free PMC article.
-
Conserved nuclear export sequences in Schizosaccharomyces pombe Mex67 and human TAP function in mRNA export by direct nuclear pore interactions.J Biol Chem. 2004 Apr 23;279(17):17434-42. doi: 10.1074/jbc.M309731200. Epub 2004 Feb 12. J Biol Chem. 2004. PMID: 14963046
-
The nuclear export signal of splicing factor Uap56p interacts with nuclear pore-associated protein Rae1p for mRNA export in Schizosaccharomyces pombe.J Biol Chem. 2007 Jun 15;282(24):17507-16. doi: 10.1074/jbc.M609727200. Epub 2007 Apr 20. J Biol Chem. 2007. PMID: 17449473
-
A novel family of nuclear transport receptors mediates the export of messenger RNA to the cytoplasm.Eur J Cell Biol. 2002 Nov;81(11):577-84. doi: 10.1078/0171-9335-00273. Eur J Cell Biol. 2002. PMID: 12498157 Review.
-
Nucleocytoplasmic transport enters the atomic age.Curr Opin Cell Biol. 2001 Jun;13(3):310-9. doi: 10.1016/s0955-0674(00)00213-1. Curr Opin Cell Biol. 2001. PMID: 11343901 Review.
Cited by
-
Mug28, a meiosis-specific protein of Schizosaccharomyces pombe, regulates spore wall formation.Mol Biol Cell. 2010 Jun 15;21(12):1955-67. doi: 10.1091/mbc.e09-12-0997. Epub 2010 Apr 21. Mol Biol Cell. 2010. PMID: 20410137 Free PMC article.
-
Global coordination of transcriptional control and mRNA decay during cellular differentiation.Mol Syst Biol. 2010 Jun 8;6:380. doi: 10.1038/msb.2010.38. Mol Syst Biol. 2010. PMID: 20531409 Free PMC article.
-
The Fission Yeast RNA-Binding Protein Meu5 Is Involved in Outer Forespore Membrane Breakdown during Spore Formation.J Fungi (Basel). 2020 Nov 13;6(4):284. doi: 10.3390/jof6040284. J Fungi (Basel). 2020. PMID: 33202882 Free PMC article.
-
The CCR4-NOT complex is implicated in the viability of aneuploid yeasts.PLoS Genet. 2012;8(6):e1002776. doi: 10.1371/journal.pgen.1002776. Epub 2012 Jun 21. PLoS Genet. 2012. PMID: 22737087 Free PMC article.
References
-
- Alfa C, Fantes P, Hyams J, Mcleod M, Warbrick E. Experiments with Fission Yeast. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1993.
-
- Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. - PubMed
-
- Berbet N, Muriel WJ, Carr AM. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene. 1992;114:59–66. - PubMed
-
- Brown JA, Bharathi A, Ghosh A, Whalen W, Fitzgerald E, Dhar R. A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+ RNA export and in the cytoskeleton. J Biol Chem. 1995;270:7411–7419. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

