Dissection of transient oxidative stress response in Saccharomyces cerevisiae by using DNA microarrays
- PMID: 12181346
- PMCID: PMC117942
- DOI: 10.1091/mbc.e02-02-0075
Dissection of transient oxidative stress response in Saccharomyces cerevisiae by using DNA microarrays
Abstract
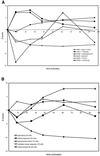
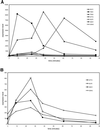
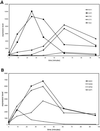
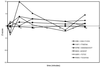
Yeast cells were grown in glucose-limited chemostat cultures and forced to switch to a new carbon source, the fatty acid oleate. Alterations in gene expression were monitored using DNA microarrays combined with bioinformatics tools, among which was included the recently developed algorithm REDUCE. Immediately after the switch to oleate, a transient and very specific stress response was observed, followed by the up-regulation of genes encoding peroxisomal enzymes required for fatty acid metabolism. The stress response included up-regulation of genes coding for enzymes to keep thioredoxin and glutathione reduced, as well as enzymes required for the detoxification of reactive oxygen species. Among the genes coding for various isoenzymes involved in these processes, only a specific subset was expressed. Not the general stress transcription factors Msn2 and Msn4, but rather the specific factor Yap1p seemed to be the main regulator of the stress response. We ascribe the initiation of the oxidative stress response to a combination of poor redox flux and fatty acid-induced uncoupling of the respiratory chain during the metabolic reprogramming phase.
Figures




References
-
- Baumgartner U, Hamilton B, Piskacek M, Ruis H, Rottensteiner H. Functional analysis of the Zn2Cys6 transcription factors Oaf1p and Pip2p. J Biol Chem. 1999;274:22208–22216. - PubMed
-
- Beck T, Hall MN. The TOR signaling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. - PubMed
-
- Beevers H. Glyoxysomes of castor bean endosperm and their relation to gluconeogenesis. Ann NY Acad Sci. 1969;168:313–324. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

