Characterization of sparsomycin resistance in Streptomyces sparsogenes
- PMID: 12183247
- PMCID: PMC127450
- DOI: 10.1128/AAC.46.9.2914-2919.2002
Characterization of sparsomycin resistance in Streptomyces sparsogenes
Abstract
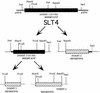
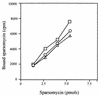
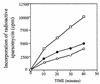
The antitumor antibiotic sparsomycin, produced by Streptomyces sparsogenes, is a universal translation inhibitor that blocks the peptide bond formation in ribosomes from all species. Sparsomycin-resistant strains were selected by transforming the sensitive Streptomyces lividans with an S. sparsogenes library. Resistance was linked to the presence of a plasmid containing an S. sparsogenes 5.9-kbp DNA insert. A restriction analysis of the insert traced down the resistance to a 3.6-kbp DNA fragment, which was sequenced. The analysis of the fragment nucleotide sequence together with the previous restriction data associate the resistance to srd, an open reading frame of 1,800 nucleotides. Ribosomes from S. sparsogenes and the S. lividans-resistant strains are equally sensitive to the inhibitor and bind the drug with similar affinity. Moreover, the drug was not modified by the resistant strains. However, resistant cells accumulated less antibiotic than the sensitive ones. In addition, membrane fractions from the resistant strains showed a higher capacity for binding the drug. The results indicate that resistance in the producer strain is not connected to either ribosome modification or drug inactivation, but it might be related to an alteration in the sparsomycin permeability barrier.
Figures




Similar articles
-
Sparsomycin Biosynthesis Highlights Unusual Module Architecture and Processing Mechanism in Non-ribosomal Peptide Synthetase.ACS Chem Biol. 2015 Aug 21;10(8):1765-9. doi: 10.1021/acschembio.5b00284. Epub 2015 Jun 9. ACS Chem Biol. 2015. PMID: 26046698
-
Direct crosslinking of the antitumor antibiotic sparsomycin, and its derivatives, to A2602 in the peptidyl transferase center of 23S-like rRNA within ribosome-tRNA complexes.Proc Natl Acad Sci U S A. 1999 Aug 3;96(16):9003-8. doi: 10.1073/pnas.96.16.9003. Proc Natl Acad Sci U S A. 1999. PMID: 10430885 Free PMC article.
-
Draft genome sequence of broad-spectrum antibiotic sparsomycin-producing Streptomyces sparsogenes ATCC 25498 from the American Type Culture Collection.J Glob Antimicrob Resist. 2017 Dec;11:159-160. doi: 10.1016/j.jgar.2017.10.011. Epub 2017 Oct 27. J Glob Antimicrob Resist. 2017. PMID: 29111481
-
Chemical and biological aspects of sparsomycin, an antibiotic from Streptomyces.Prog Med Chem. 1986;23:219-68. doi: 10.1016/s0079-6468(08)70344-8. Prog Med Chem. 1986. PMID: 3310108 Review. No abstract available.
-
Chemical, biochemical and genetic endeavours characterizing the interaction of sparsomycin with the ribosome.Biochimie. 1991 Jul-Aug;73(7-8):1137-43. doi: 10.1016/0300-9084(91)90157-v. Biochimie. 1991. PMID: 1720666 Review.
References
-
- Cundliffe, E. 1989. How antibiotic-producing organisms avoid suicide. Annu. Rev. Microbiol. 43:207-233. - PubMed
-
- Cundliffe, E., and J. Thompson. 1979. Ribose methylation and resistance to thiostrepton. Nature 278:859-861. - PubMed
-
- Fiebig, H. H., D. P. Berger, K. Kopping, H. C. Ottenheijm, and Z. Zylicz. 1990. In vitro and in vivo anticancer activity of mitozolomide and sparsomycin in human tumor xenografts, murine tumors and human bone marrow. J. Cancer Res. Clin. Oncol. 116:550-556. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

