Saccharomyces forkhead protein Fkh1 regulates donor preference during mating-type switching through the recombination enhancer
- PMID: 12183363
- PMCID: PMC186439
- DOI: 10.1101/gad.994902
Saccharomyces forkhead protein Fkh1 regulates donor preference during mating-type switching through the recombination enhancer
Abstract
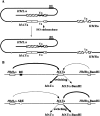
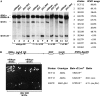
Saccharomyces mating-type switching results from replacement by gene conversion of the MAT locus with sequences copied from one of two unexpressed donor loci, HML or HMR. MATa cells recombine with HMLalpha approximately 90% of the time, whereas MATalpha cells choose HMRa 80%-90% of the time. HML preference in MATa is controlled by the cis-acting recombination enhancer (RE) that regulates recombination along the entire left arm of chromosome III. Comparison of RE sequences between S. cerevisiae, S. carlsbergensis, and S. bayanus defines four highly conserved regions (A, B, C, and D) within a 270-bp minimum RE. An adjacent E region enhances RE activity. Multimers of region A, D, or E are sufficient to promote selective use of HML. Regions A, D, and E each bind in vivo the transcription activator forkhead proteins Fkh1p and Fkh2p and their associated Ndd1p, although there are no adjacent open reading frames (ORFs). Deletion of FKH1 significantly reduces MATa's use of HML, as does mutation of the Fkh1/Fkh2-binding sites in a multimer of region A. We conclude that Fkh1p regulates MATa donor preference through direct interaction with RE.
Figures






References
-
- Bon E, Neuveglise C, Casaregola S, Artiguenave F, Wincker P, Aigle M, Durrens P. Genomic exploration of the hemiascomycetous yeasts: 5. Saccharomyces bayanus var. uvarum. FEBS Lett. 2000;487:37–41. - PubMed
-
- Chen DC, Yang BC, Kuo TT. One-step transformation of yeast in stationary phase. Curr Genet. 1992;21:83–84. - PubMed
-
- Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR. Ploidy regulation of gene expression. Science. 1999;285:251–254. - PubMed
-
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. - PubMed
-
- Gietz RD, Triggs-Raine B, Robbins A, Graham KC, Woods RA. Identification of proteins that interact with a protein of interest: Applications of the yeast two-hybrid system. Mol Cell Biochem. 1997;172:67–79. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
