DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response
- PMID: 12183369
- PMCID: PMC186436
- DOI: 10.1101/gad.1008902
DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response
Abstract
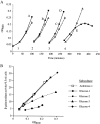
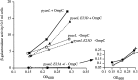
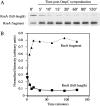
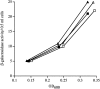
All cells have stress response pathways that maintain homeostasis in each cellular compartment. In the Gram-negative bacterium Escherichia coli, the sigma(E) pathway responds to protein misfolding in the envelope. The stress signal is transduced across the inner membrane to the cytoplasm via the inner membrane protein RseA, the anti-sigma factor that inhibits the transcriptional activity of sigma(E). Stress-induced activation of the pathway requires the regulated proteolysis of RseA. In this report we show that RseA is degraded by sequential proteolytic events controlled by the inner membrane-anchored protease DegS and the membrane-embedded metalloprotease YaeL, an ortholog of mammalian Site-2 protease (S2P). This is consistent with the mechanism of activation of ATF6, the mammalian unfolded protein response transcription factor by Site-1 protease and S2P. Thus, mammalian and bacterial cells employ a conserved proteolytic mechanism to activate membrane-associated transcription factors that initiate intercompartmental cellular stress responses.
Figures



References
-
- Alba BM, Zhong HJ, Pelayo JC, Gross CA. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide σEactivity. Mol Microbiol. 2001;40:1323–1333. - PubMed
-
- Bartolomé B, Jubete Y, Martinez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. - PubMed
-
- Beebe KD, Shin J, Peng J, Chaudhury C, Khera J, Pei D. Substrate recognition through a PDZ domain in tail-specific protease. Biochemistry. 2000;39:3149–3155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
