Molecular genetic analyses of mating pheromones reveal intervariety mating or hybridization in Cryptococcus neoformans
- PMID: 12183574
- PMCID: PMC128272
- DOI: 10.1128/IAI.70.9.5225-5235.2002
Molecular genetic analyses of mating pheromones reveal intervariety mating or hybridization in Cryptococcus neoformans
Abstract
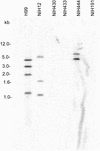
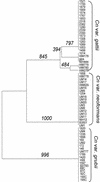
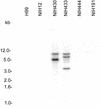
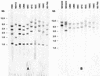
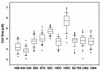
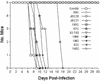
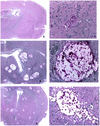
The sexual mating of the pathogenic yeast Cryptococcus neoformans is important for pathogenesis studies because the fungal virulence is linked to the alpha mating type (MAT(alpha)). We characterized C. neoformans mating pheromones (MF(alpha) 1 and MFa1) from 122 strains to understand intervariety hybridization or mating and intervariety virulence. MF(alpha) 1 in three C. neoformans varieties showed (a) specific nucleotide polymorphisms, (b) different copy numbers and chromosomal localizations, and (c) unique deduced amino acids in two geographic populations of C. neoformans var. gattii. MF(alpha) 1 of different varieties cross-hybridized in Southern hybridizations. Their phylogenetic analyses showed purifying selection (neutral evolution). These observations suggested that MAT(alpha) strains from any of the three C. neoformans varieties could mate or hybridize in nature with MATa strains of C. neoformans var. neoformans. A few serotype A/D diploid strains provided evidence for mating or hybridization, while a majority of A/D strains tested positive for haploid MF(alpha) 1 identical to that of C. neoformans var. grubii. MF(alpha) 1 sequence and copy numbers in diploids were identical to those of C. neoformans var. grubii, while their MFa1 sequences were identical to those of C. neoformans var. neoformans; thus, these strains were hybrids. The mice survival curves and histological lesions revealed A/D diploids to be highly pathogenic, with pathogenicity levels similar to that of the C. neoformans var. grubii type strain and unlike the low pathogenicity levels of C. neoformans var. neoformans strains. In contrast to MF(alpha) 1 in three varieties, MFa1 amplicons and hybridization signals could be obtained only from two C. neoformans var. neoformans reference strains and eight A/D diploids. This suggested that a yet undiscovered MFa pheromone(s) in C. neoformans var. gattii and C. neoformans var. grubii is unrelated to, highly divergent from, or rarer than that in C. neoformans var. neoformans. These observations could form the basis for future studies on the role of intervariety mating in C. neoformans biology and virulence.
Figures

Similar articles
-
Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates.Infect Immun. 2003 Sep;71(9):4831-41. doi: 10.1128/IAI.71.9.4831-4841.2003. Infect Immun. 2003. PMID: 12933823 Free PMC article.
-
Many globally isolated AD hybrid strains of Cryptococcus neoformans originated in Africa.PLoS Pathog. 2007 Aug 17;3(8):e114. doi: 10.1371/journal.ppat.0030114. PLoS Pathog. 2007. PMID: 17708680 Free PMC article.
-
Molecular and genetic characterization of a serotype A MATa Cryptococcus neoformans isolate.Microbiology (Reading). 2003 Jan;149(Pt 1):131-42. doi: 10.1099/mic.0.25921-0. Microbiology (Reading). 2003. PMID: 12576587
-
[Mating types, sexual reproduction and ploidy in fungi: effects on virulence].Mikrobiyol Bul. 2009 Jul;43(3):507-13. Mikrobiyol Bul. 2009. PMID: 19795629 Review. Turkish.
-
Molecular analyses of the serotype of Cryptococcus neoformans.Nihon Ishinkin Gakkai Zasshi. 2001;42(2):69-74. doi: 10.3314/jjmm.42.69. Nihon Ishinkin Gakkai Zasshi. 2001. PMID: 11331466 Review.
Cited by
-
Extracellular fibrils of pathogenic yeast Cryptococcus gattii are important for ecological niche, murine virulence and human neutrophil interactions.PLoS One. 2010 Jun 7;5(6):e10978. doi: 10.1371/journal.pone.0010978. PLoS One. 2010. PMID: 20539754 Free PMC article.
-
Cryptococcus gattii, no longer an accidental pathogen?Curr Fungal Infect Rep. 2012 Dec;6(4):245-256. doi: 10.1007/s12281-012-0111-0. Curr Fungal Infect Rep. 2012. PMID: 23243480 Free PMC article.
-
Genetic Diversity and Genomic Plasticity of Cryptococcus neoformans AD Hybrid Strains.G3 (Bethesda). 2012 Jan;2(1):83-97. doi: 10.1534/g3.111.001255. Epub 2012 Jan 1. G3 (Bethesda). 2012. PMID: 22384385 Free PMC article.
-
Microreview: capsule-associated genes of Cryptococcus neoformans.Mycopathologia. 2007 Jan;163(1):1-8. doi: 10.1007/s11046-006-0083-0. Mycopathologia. 2007. PMID: 17216326 Review.
-
Heterozygosis and pathogenicity of Cryptococcus neoformans AD-hybrid isolates.Mycopathologia. 2012 Jun;173(5-6):347-57. doi: 10.1007/s11046-011-9467-x. Epub 2011 Sep 16. Mycopathologia. 2012. PMID: 21922248
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
-
- Boekhout, T., B. Theelen, M. Diaz, J. W. Fell, W. C. J. Hop, E. C. A. Abeln, F. Dromer, and W. Meyer. 2001. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology 147:891-907. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

