Identification and disruption of the gene encoding the third member of the low-molecular-mass rhoptry complex in Plasmodium falciparum
- PMID: 12183575
- PMCID: PMC128283
- DOI: 10.1128/IAI.70.9.5236-5245.2002
Identification and disruption of the gene encoding the third member of the low-molecular-mass rhoptry complex in Plasmodium falciparum
Abstract
The low-molecular-mass rhoptry complex of Plasmodium falciparum consists of three proteins, rhoptry-associated protein 1 (RAP1), RAP2, and RAP3. The genes encoding RAP1 and RAP2 are known; however, the RAP3 gene has not been identified. In this study we identify the RAP3 gene from the P. falciparum genome database and show that this protein is part of the low-molecular-mass rhoptry complex. Disruption of RAP3 demonstrated that it is not essential for merozoite invasion, probably because RAP2 can complement the loss of RAP3. RAP3 has homology with RAP2, and the genes are encoded on chromosome 5 in a head-to-tail fashion. Analysis of the genome databases has identified homologous genes in all Plasmodium spp., suggesting that this protein plays a role in merozoite invasion. The region surrounding the RAP3 homologue in the Plasmodium yoelii genome is syntenic with the same region in P. falciparum; however, there is a single gene. Phylogenetic comparison of the RAP2/3 protein family from Plasmodium spp. suggests that the RAP2/3 duplication occurred after divergence of these parasite species.
Figures

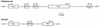
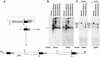




References
-
- Anders, R. F. 1997. Vaccines against asexual blood-stages of Plasmodium falciparum, p. 1037-1057. In M. M. Levine, G. Woodrow, G. Cobon, J. Kaper, and G. Garrett (ed.), New generation vaccines, 2nd ed., vol. 63. Marcel Dekker, Inc., New York, N.Y.
-
- Bahl, A., B. Brunk, R. L. Coppel, J. Crabtree, S. J. Diskin, M. J. Fraunholz, G. R. Grant, D. Gupta, R. L. Huestis, J. C. Kissinger, P. Labo, L. Li, S. K. McWeeney, A. J. Milgram, D. S. Roos, J. Schug, and C. J. Stoeckert, Jr. 2002. PlasmoDB: the Plasmodium genome resource. An integrated database providing tools for accessing, analyzing and mapping expression and sequence data (both finished and unfinished). Nucleic Acids Res. 30:87-90. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

