Profound effect of normalization on detection of differentially expressed genes in oligonucleotide microarray data analysis
- PMID: 12184807
- PMCID: PMC126238
- DOI: 10.1186/gb-2002-3-7-research0033
Profound effect of normalization on detection of differentially expressed genes in oligonucleotide microarray data analysis
Abstract
Background: Oligonucleotide microarrays measure the relative transcript abundance of thousands of mRNAs in parallel. A large number of procedures for normalization and detection of differentially expressed genes have been proposed. However, the relative impact of these methods on the detection of differentially expressed genes remains to be determined.
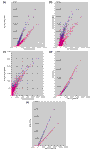
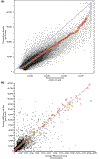
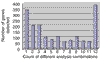
Results: We have employed four different normalization methods and all possible combinations with three different statistical algorithms for detection of differentially expressed genes on a prototype dataset. The number of genes detected as differentially expressed differs by a factor of about three. Analysis of lists of genes detected as differentially expressed, and rank correlation coefficients for probability of differential expression shows that a high concordance between different methods can only be achieved by using the same normalization procedure.
Conclusions: Normalization has a profound influence of detection of differentially expressed genes. This influence is higher than that of three subsequent statistical analysis procedures examined. Algorithms incorporating more array-derived information than gene-expression values alone are urgently needed.
Figures

References
-
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. - PubMed
-
- Wodicka L, Dong H, Mittmann M, Ho MH, Lockhart DJ. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. - PubMed
-
- Fambrough D, McClure K, Kazlauskas A, Lander ES. Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell. 1999;97:727–741. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

