Isolation of high-affinity GTP aptamers from partially structured RNA libraries
- PMID: 12185247
- PMCID: PMC129318
- DOI: 10.1073/pnas.182095699
Isolation of high-affinity GTP aptamers from partially structured RNA libraries
Abstract
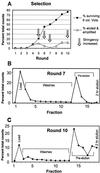
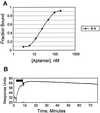
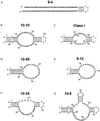
Aptamers, RNA sequences that bind to target ligands, are typically isolated by in vitro selection from RNA libraries containing completely random sequences. To see whether higher-affinity aptamers can be isolated from partially structured RNA libraries, we selected for aptamers that bind GTP, starting from a mixture of fully random and partially structured libraries. Because stem-loops are common motifs in previously characterized aptamers, we designed the partially structured library to contain a centrally located stable stem-loop. We used an off-rate selection protocol designed to maximize the enrichment of high-affinity aptamers. The selection produced a surprisingly large number of distinct sequence motifs and secondary structures, including seven different aptamers with K(d)s ranging from 500 to 25 nanomolar. The engineered stem-loop was present in the three highest affinity aptamers, and in 12 of 13 independent isolates with a single consensus sequence, suggesting that its inclusion increased the abundance of high-affinity aptamers in the starting pool.
Figures

References
-
- Ellington A D, Szostak J W. Nature (London) 1990;346:818–822. - PubMed
-
- Tuerk C, Gold L. Science. 1990;249:505–510. - PubMed
-
- Wilson D S, Szostak J W. Annu Rev Biochem. 1999;68:611–647. - PubMed
-
- Hesselberth J, Robertson M P, Jhaveri S, Ellington A D. J Biotechnol. 2000;74:15–25. - PubMed
-
- Famulok M. Curr Opin Struct Biol. 1999;9:324–329. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

