Expression profiling of Drosophila imaginal discs
- PMID: 12186645
- PMCID: PMC126232
- DOI: 10.1186/gb-2002-3-8-research0038
Expression profiling of Drosophila imaginal discs
Abstract
Background: In the Drosophila larva, imaginal discs are programmed to produce adult structures at metamorphosis. Although their fate is precisely determined, these organs remain largely undifferentiated in the larva. To identify genes that establish and express the different states of determination in discs and larval tissues, we used DNA microarrays to analyze mRNAs isolated from single imaginal discs.
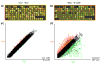
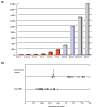
Results: Linear amplification protocols were used to generate hybridization probes for microarray analysis from poly(A)+ RNA from single imaginal discs containing between 10,000 and 60,000 cells. Probe reproducibility and degree of representation were tested using microarrays with approximately 6,000 different cDNAs. Hybridizations with probes that had been prepared separately from the same starting RNA pool had a correlation coefficient of 0.97. Expression-profile comparisons of the left and right wing imaginal discs from the same larva correlated with a coefficient of 0.99, indicating a high degree of reproducibility of independent amplifications. Using this method, we identified genes with preferential expression in the different imaginal discs using pairwise comparisons of discs and larval organs. Whereas disc-to-disc comparisons revealed only moderate differences, profiles differed substantially between imaginal discs and larval tissues, such as larval endodermal midgut and mesodermal fat body.
Conclusions: The combination of linear RNA amplification and DNA microarray hybridization allowed us to determine the expression profiles of individual imaginal discs and larval tissues and to identify genes expressed in tissue-specific patterns. These methods should be widely applicable to comparisons of expression profiles for tissues or parts of tissues that are available only in small amounts.
Figures




References
-
- Struhl G. A homoeotic mutation transforming leg to antenna in Drosophila. Nature. 1981;292:635–638. - PubMed
-
- Moses K, Ellis MC, Rubin GM. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature. 1989;340:531–536. - PubMed
-
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

