BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data
- PMID: 12186655
- PMCID: PMC139402
- DOI: 10.1186/gb-2002-3-8-software0003
BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data
Abstract
The microarray technique requires the organization and analysis of vast amounts of data. These data include information about the samples hybridized, the hybridization images and their extracted data matrices, and information about the physical array, the features and reporter molecules. We present a web-based customizable bioinformatics solution called BioArray Software Environment (BASE) for the management and analysis of all areas of microarray experimentation. All software necessary to run a local server is freely available.
Figures
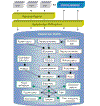


References
-
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

